Main Conclusion
Time series analysis was conducted on Air Quality data and built predictive models for the variables 'PT08.S1', 'PT08.S2', 'PT08.S3', 'PT08.S4', and 'PT08.S5'. To systematically understand the historical characteristics of the time series, the analysis was divied into univariate analysis and multivariate analysis. Based on these analyses, several models was constructed to estimate future time series values, and its performance was evaluated. In conclusion, the predictor variables exhibit a long-term equilibrium relationship, showing a generally mutual influence among them. Among these variables, only PT08.S1 was found not to Granger-cause the other variables.
Summary for Results by Analysis Procedures
Step01. Data Extraction
- Missing Value & Duplication Inspection: The given data consists of a total of 9,357 entries with 13 columns during 2004-03-10 18:00:00 ~ 2005-04-04 14:00:00. Among these, the highest number of missing values was identified with 8,443 cases. The remaining columns exhibit missing values of either 366 cases or 1,639 cases. In terms of duplication, 'CO(GT)', 'NO2(GT)', 'C6H6(GT)', 'NMHC(GT)' columns exhibit high levels of duplication in that order.
- Data Interpolation through Gaussian Process: The missing values were filled in using Gaussian interpolation. All columns except for NMHC(GT) satisfy null integrity. And only NMHC(GT) has 8,126 missing values. The reason Gaussian interpolation was used is to ensure the exisiting stattistical statbility, thereby preserving the time series characteristics.
Step02. Exploratory Data Analysis
- Descriptive statistics: The time series unit is in hours, and it has verified that the count of weekly data is evenly distributed as 24*7, 168. Additionally, the time series characteristics of montly auto-correlation, heteroscedasticity, normality and stationary are described. Lastly, the cycle subseries of the time series are visualized hourly, monthly.
- Weekly description: count, max, min, mean, standard deviation
- Monthly description: auto-correlation, heteroscedasticity, normality, stationary
- Cycle subseries description: visualization about temporary seasonal series
- Exploration for time series process: The fundamental approach to time series analysis involves distinguishing between deterministic processes and stochastics processes. Buling upon this, the robust nature of STL(Seasonal and Trend decomposition using Loess) is utlized to explore deterministic process. While UCM(Unobserved Component Model) is employed to investigate stochastic processes. Ultimately, this enables examination of the processes related to trend and seasonality.
- Deterministic process of time series features: STL
- Stochastic process of time series features: UCM
Step03. Univariate Time Series Characteristic Analysis
- Stationary Analysis: Stationary refers to the property where the statistical characteristics of a time series remain consistent over time regardless of the lag. If stationary is not maintained, it become challengging to define the characteristics of the time series. Therefore, it's crucial initially to assess the stationary of a time series. To account for both difference and trend stationary, both the Augumented Dickey-Fuller (ADF) test and the Kwiatkowski-Phillips-Schmidt-Shin (KPSS) test are utilized.
- Difference Stationary: ADF
- Trend Stationary: KPSS
- Auto-Correlation/Covariance Analysis: The autocorrelation/autocovariance structure of a time series play a crucial role in time series analysis, as it ultimately determines the trend and seasonality of the time series. To assess this structure while maintaining the stationary property of the time series, the autocorrelation and autocovariance structure were visualized at various time intervals, including per time unit, monthly, quarterly, yearly.
- ACF and PACF: Yule-Walker Method
- Auto-Correlation/Covariance Structure: sample covariance method; ordinary least squares
- Volatility Analysis: The statistical properties of stochastic process are primarily determined by their first and second moments. Typically, when analyzing autocorrelation structure, it is assumed that the population parameters of the first and second moments remain constant. Conversly, volatility analysis seeks to understand conditional volatility by considering time dependency in the second moment.
- GJR-GARCH(Generalized AutoRegressive Conditional Heteroskedasticity)
- Regime Analysis: Regime analysis can understood as an extension of volatility analysis, where the time dependency of the second moment is defined by transition probabliities between regimes. It involves observing regime-specific durations and determining transition probabilities between regimes. In conclusion, it has been confirmed that there is no need to consider volatility for the pollutant variables 'PT08.S1(CO)', 'PT08.S2(NMHC)', 'PT08.S3(NOx)', 'PT08.S4(NO2)', 'PT08.S5(O3)'.
- MSM(Markov Switching Model)
Step04. Multivariate Time Series Characteristic Analysis
- Cross-Covariance Analysis: To understand the covariance patterns on an hourly, weekly, monthly, monthly on year, analysis of covariance (ANCOVA) is conducted. This is done to identify key date factors that influence the time series characteristics.
- ANCOVA: Analysis of Covariance
- Cointegration Analysis: To avoid spurious regression issues, cointegration relationship was explored in the time series. By johansen cointegration test, the cointegration rank has been determined as 4.
- Bivariate Cointegration Analysis: Augmented Engle-Granger Method
- Multivariate Cointegration Analysis: Johansen Cointegration Test
- Principal Component Analysis(PCA): The first principal component to view the random variable for the given data from a different perspective is a set of the pollutants variables including 'NMHC(GT)'.; ['PT08.S1', 'PT08.S2', 'PT08.S3', 'PT08.S4', 'PT08.S5']
- PCA for Stochastic Time Series Compoents
- Dynamic Factor Analysis(DFA): As a key factor, along with the first principal component about results in PCA, 'CO(GT)', 'C6H6(GT)', 'NOx(GT)', 'NO2(GT)' are loaded into the common loadings of the first factor while 'T', 'RH' and 'AH' remain as unique loadings.
Step05. Predictive Modeling and Evaluation
- Time Series Model Design: All modeling was conducted under the assumption that time series exhibit deterministic seasonality and stochastic trends. The purpose is to build and evaluate a time series model for stochastic trends with removed seasonality.
- Overall Model Specification
- Base Preprocessing for Forecasting Pollutants Variables
- Time Series Model Evaluation and Forecast: The time series models were constructed into baseline and improved models, and the results of h-step forecast evaluations were presented.
- Baseline Explanation Model
- Naive Forecast, ETS(AAN)
- Improvement Model
- Predictor variable: pollutants: 'PT08.S1', 'PT08.S2', 'PT08.S3', 'PT08.S4', 'PT08.S5'
- Short-Term Forecast
- Univariate: ARDL, SARIMAX, SVM
- Multivariate: VARMAX[Reduced, Structural-Orthogonal Residual]
- Baseline Explanation Model
- Impulse Response Analysis: Through Impulse Response Analysis (IRA), the potential tapering-off effects that can occur if there are long-term equilibrium relationships was explored to understand the characteristics of endogenous variables in the time series model. As a result, PT08.S1(CO) was found to not granger cause other pollutants.
- IR Analysis Model
- Univariate Orthogonalized IRF: SARIMAX
- Multivariate Orthogonalized IRF: VAR/VECM
- Granger Causality Analysis: VAR/VECM
- IR Analysis Model
Step06. Summary for Results by Analysis Procedures
Step07. Main Conclusion
Predictive Modeling and Evaluation
Impulse Response and Granger Causality Analysis
Univariate IRF: SARIMAX
- endogenous variables: PT08.S1(CO), PT08.S2(NMHC), PT08.S3(NOx), PT08.S4(NO2), PT08.S5(O3), CO(GT), NMHC(GT), C6H6(GT), NOx(GT), NO2(GT), T, RH, AH
- autoregressive order: 2
- [IRF Train Period]: 2004-03-10 18:00:00 ~ 2004-05-01 00:00:00, 1231

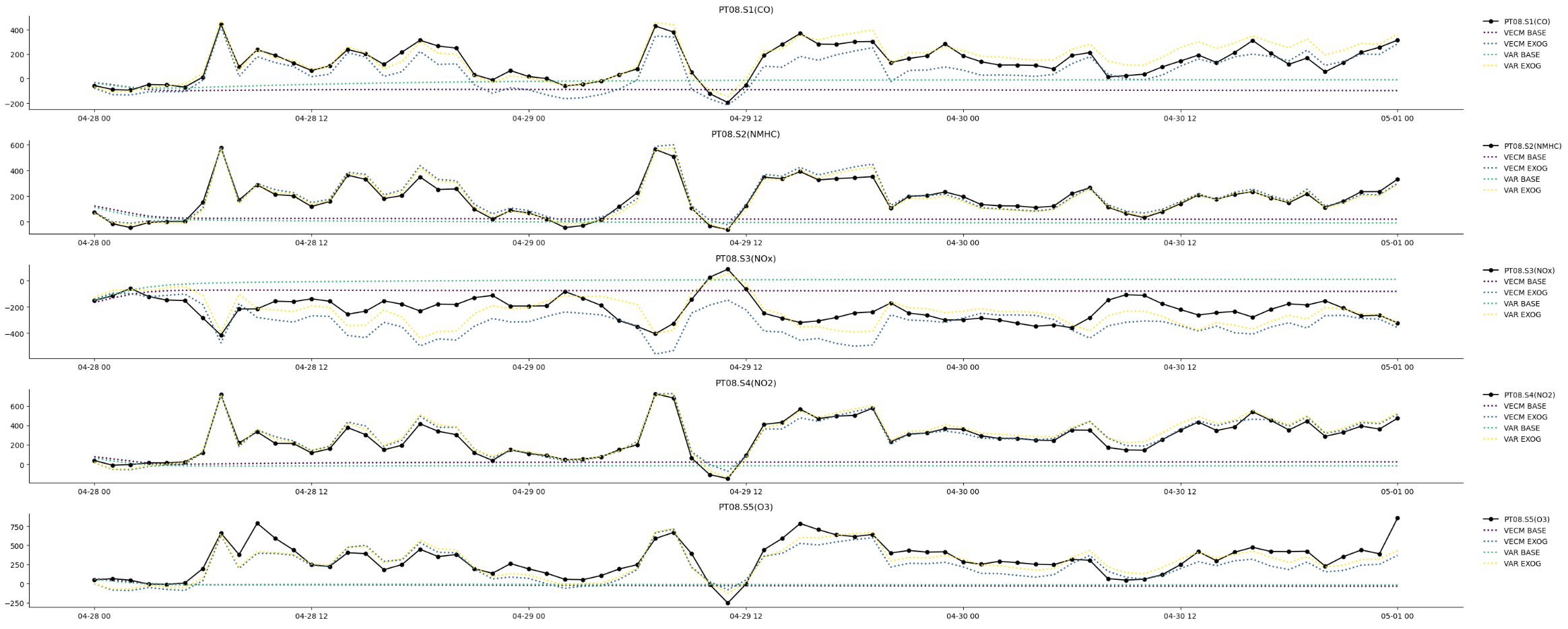
Multivariate IRF: VAR/VECM
- endogenous variables: PT08.S1(CO), PT08.S2(NMHC), PT08.S3(NOx), PT08.S4(NO2), PT08.S5(O3)
- cointegration exogenous variables: CO(GT), NMHC(GT), C6H6(GT), NOx(GT), NO2(GT)
- exogenous variables: T, RH, AH
- cointegration rank: 4
- autoregressive order: 2
- [Train Period]: 2004-03-10 18:00:00 ~ 2004-04-27 23:00:00, 1158
- [Test Period]: 2004-04-28 00:00:00 ~ 2004-05-01 00:00:00, 73
- [IRF Train Period]: 2004-03-10 18:00:00 ~ 2004-05-01 00:00:00, 1231

Granger Causality Analysis
PT08.S1(CO) does not Granger-cause PT08.S2(NMHC)
PT08.S2(NMHC) do Granger-cause PT08.S1(CO)
PT08.S1(CO) does not Granger-cause PT08.S3(NOx)
PT08.S3(NOx) do Granger-cause PT08.S1(CO)
PT08.S1(CO) does not Granger-cause PT08.S4(NO2)
PT08.S4(NO2) do Granger-cause PT08.S1(CO)
PT08.S1(CO) does not Granger-cause PT08.S5(O3)
PT08.S5(O3) do Granger-cause PT08.S1(CO)
PT08.S4(NO2) does not Granger-cause PT08.S3(NOx)
PT08.S3(NOx) do Granger-cause PT08.S4(NO2)
PT08.S5(O3) does not Granger-cause PT08.S1(CO)
PT08.S1(CO) do Granger-cause PT08.S5(O3)
PT08.S5(O3) does not Granger-cause PT08.S4(NO2)
PT08.S4(NO2) do Granger-cause PT08.S5(O3)
| By cause | Effect | Causality type | Test statistic | Critical value | p-value | df | H0: Null hypothesis | Test result | Granger Causality | Instantaneous Causality | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | PT08.S1(CO) | PT08.S1(CO) | Granger causality F-test | 242.976132 | 2.606392 | 8.685386e-149 | (3, 5990) | PT08.S1(CO) does not Granger-cause PT08.S1(CO) | reject H_0 at 5% significance level. | True | - |
| 6 | PT08.S2(NMHC) | PT08.S1(CO) | Granger causality F-test | 38.390892 | 2.606392 | 1.446181e-24 | (3, 5990) | PT08.S2(NMHC) does not Granger-cause PT08.S1(CO) | reject H_0 at 5% significance level. | True | - |
| 7 | PT08.S3(NOx) | PT08.S1(CO) | Granger causality F-test | 11.514925 | 2.606392 | 1.593106e-07 | (3, 5990) | PT08.S3(NOx) does not Granger-cause PT08.S1(CO) | reject H_0 at 5% significance level. | True | - |
| 8 | PT08.S4(NO2) | PT08.S1(CO) | Granger causality F-test | 29.948117 | 2.606392 | 3.278482e-19 | (3, 5990) | PT08.S4(NO2) does not Granger-cause PT08.S1(CO) | reject H_0 at 5% significance level. | True | - |
| 9 | PT08.S5(O3) | PT08.S1(CO) | Granger causality F-test | 2.075985 | 2.606392 | 1.011516e-01 | (3, 5990) | PT08.S5(O3) does not Granger-cause PT08.S1(CO) | fail to reject H_0 at 5% significance level. | False | - |
| 5 | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\', \'PT08.S4(NO2)\', \'PT08.S5(O3)\'] | PT08.S1(CO) | Granger causality F-test | 76.184574 | 1.753786 | 1.463578e-174 | (12, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\', \'PT08.S4(NO2)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S1(CO) | reject H_0 at 5% significance level. | True | - |
| 16 | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] | PT08.S1(CO) | Granger causality F-test | 94.129400 | 1.881443 | 7.445732e-165 | (9, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] do not Granger-cause PT08.S1(CO) | reject H_0 at 5% significance level. | True | - |
| 17 | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\', \'PT08.S5(O3)\'] | PT08.S1(CO) | Granger causality F-test | 16.083500 | 1.881443 | 2.346265e-26 | (9, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S1(CO) | reject H_0 at 5% significance level. | True | - |
| 10 | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\'] | PT08.S1(CO) | Granger causality F-test | 23.297272 | 2.100103 | 2.375558e-27 | (6, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\'] do not Granger-cause PT08.S1(CO) | reject H_0 at 5% significance level. | True | - |
| 18 | [\'PT08.S2(NMHC)\', \'PT08.S4(NO2)\', \'PT08.S5(O3)\'] | PT08.S1(CO) | Granger causality F-test | 84.093331 | 1.881443 | 1.045150e-147 | (9, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S4(NO2)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S1(CO) | reject H_0 at 5% significance level. | True | - |
| 11 | [\'PT08.S2(NMHC)\', \'PT08.S4(NO2)\'] | PT08.S1(CO) | Granger causality F-test | 122.328522 | 2.100103 | 2.424444e-146 | (6, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S4(NO2)\'] do not Granger-cause PT08.S1(CO) | reject H_0 at 5% significance level. | True | - |
| 12 | [\'PT08.S2(NMHC)\', \'PT08.S5(O3)\'] | PT08.S1(CO) | Granger causality F-test | 19.495824 | 2.100103 | 1.190212e-22 | (6, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S1(CO) | reject H_0 at 5% significance level. | True | - |
| 19 | [\'PT08.S3(NOx)\', \'PT08.S4(NO2)\', \'PT08.S5(O3)\'] | PT08.S1(CO) | Granger causality F-test | 16.583265 | 1.881443 | 2.896168e-27 | (9, 5990) | [\'PT08.S3(NOx)\', \'PT08.S4(NO2)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S1(CO) | reject H_0 at 5% significance level. | True | - |
| 13 | [\'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] | PT08.S1(CO) | Granger causality F-test | 23.319128 | 2.100103 | 2.232157e-27 | (6, 5990) | [\'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] do not Granger-cause PT08.S1(CO) | reject H_0 at 5% significance level. | True | - |
| 14 | [\'PT08.S3(NOx)\', \'PT08.S5(O3)\'] | PT08.S1(CO) | Granger causality F-test | 7.744475 | 2.100103 | 2.576535e-08 | (6, 5990) | [\'PT08.S3(NOx)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S1(CO) | reject H_0 at 5% significance level. | True | - |
| 15 | [\'PT08.S4(NO2)\', \'PT08.S5(O3)\'] | PT08.S1(CO) | Granger causality F-test | 16.021190 | 2.100103 | 2.286888e-18 | (6, 5990) | [\'PT08.S4(NO2)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S1(CO) | reject H_0 at 5% significance level. | True | - |
| 21 | PT08.S1(CO) | PT08.S2(NMHC) | Granger causality F-test | 1.322332 | 2.606392 | 2.651576e-01 | (3, 5990) | PT08.S1(CO) does not Granger-cause PT08.S2(NMHC) | fail to reject H_0 at 5% significance level. | False | - |
| 1 | PT08.S2(NMHC) | PT08.S2(NMHC) | Granger causality F-test | 0.299041 | 2.606392 | 8.261201e-01 | (3, 5990) | PT08.S2(NMHC) does not Granger-cause PT08.S2(NMHC) | fail to reject H_0 at 5% significance level. | False | - |
| 22 | PT08.S3(NOx) | PT08.S2(NMHC) | Granger causality F-test | 142.757856 | 2.606392 | 2.396742e-89 | (3, 5990) | PT08.S3(NOx) does not Granger-cause PT08.S2(NMHC) | reject H_0 at 5% significance level. | True | - |
| 23 | PT08.S4(NO2) | PT08.S2(NMHC) | Granger causality F-test | 6.902389 | 2.606392 | 1.230777e-04 | (3, 5990) | PT08.S4(NO2) does not Granger-cause PT08.S2(NMHC) | reject H_0 at 5% significance level. | True | - |
| 24 | PT08.S5(O3) | PT08.S2(NMHC) | Granger causality F-test | 3.178949 | 2.606392 | 2.301372e-02 | (3, 5990) | PT08.S5(O3) does not Granger-cause PT08.S2(NMHC) | reject H_0 at 5% significance level. | True | - |
| 20 | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\', \'PT08.S4(NO2)\', \'PT08.S5(O3)\'] | PT08.S2(NMHC) | Granger causality F-test | 49.673247 | 1.753786 | 5.027458e-114 | (12, 5990) | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\', \'PT08.S4(NO2)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S2(NMHC) | reject H_0 at 5% significance level. | True | - |
| 31 | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] | PT08.S2(NMHC) | Granger causality F-test | 64.258682 | 1.881443 | 3.394356e-113 | (9, 5990) | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] do not Granger-cause PT08.S2(NMHC) | reject H_0 at 5% significance level. | True | - |
| 32 | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\', \'PT08.S5(O3)\'] | PT08.S2(NMHC) | Granger causality F-test | 50.931122 | 1.881443 | 1.511647e-89 | (9, 5990) | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S2(NMHC) | reject H_0 at 5% significance level. | True | - |
| 25 | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\'] | PT08.S2(NMHC) | Granger causality F-test | 72.962868 | 2.100103 | 3.747497e-88 | (6, 5990) | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\'] do not Granger-cause PT08.S2(NMHC) | reject H_0 at 5% significance level. | True | - |
| 33 | [\'PT08.S1(CO)\', \'PT08.S4(NO2)\', \'PT08.S5(O3)\'] | PT08.S2(NMHC) | Granger causality F-test | 4.553057 | 1.881443 | 5.300025e-06 | (9, 5990) | [\'PT08.S1(CO)\', \'PT08.S4(NO2)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S2(NMHC) | reject H_0 at 5% significance level. | True | - |
| 26 | [\'PT08.S1(CO)\', \'PT08.S4(NO2)\'] | PT08.S2(NMHC) | Granger causality F-test | 3.862469 | 2.100103 | 7.516101e-04 | (6, 5990) | [\'PT08.S1(CO)\', \'PT08.S4(NO2)\'] do not Granger-cause PT08.S2(NMHC) | reject H_0 at 5% significance level. | True | - |
| 27 | [\'PT08.S1(CO)\', \'PT08.S5(O3)\'] | PT08.S2(NMHC) | Granger causality F-test | 3.013720 | 2.100103 | 6.081282e-03 | (6, 5990) | [\'PT08.S1(CO)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S2(NMHC) | reject H_0 at 5% significance level. | True | - |
| 34 | [\'PT08.S3(NOx)\', \'PT08.S4(NO2)\', \'PT08.S5(O3)\'] | PT08.S2(NMHC) | Granger causality F-test | 61.976621 | 1.881443 | 3.583655e-109 | (9, 5990) | [\'PT08.S3(NOx)\', \'PT08.S4(NO2)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S2(NMHC) | reject H_0 at 5% significance level. | True | - |
| 28 | [\'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] | PT08.S2(NMHC) | Granger causality F-test | 90.150101 | 2.100103 | 1.098180e-108 | (6, 5990) | [\'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] do not Granger-cause PT08.S2(NMHC) | reject H_0 at 5% significance level. | True | - |
| 29 | [\'PT08.S3(NOx)\', \'PT08.S5(O3)\'] | PT08.S2(NMHC) | Granger causality F-test | 73.899580 | 2.100103 | 2.800282e-89 | (6, 5990) | [\'PT08.S3(NOx)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S2(NMHC) | reject H_0 at 5% significance level. | True | - |
| 30 | [\'PT08.S4(NO2)\', \'PT08.S5(O3)\'] | PT08.S2(NMHC) | Granger causality F-test | 5.246220 | 2.100103 | 2.120826e-05 | (6, 5990) | [\'PT08.S4(NO2)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S2(NMHC) | reject H_0 at 5% significance level. | True | - |
| 36 | PT08.S1(CO) | PT08.S3(NOx) | Granger causality F-test | 0.552846 | 2.606392 | 6.462112e-01 | (3, 5990) | PT08.S1(CO) does not Granger-cause PT08.S3(NOx) | fail to reject H_0 at 5% significance level. | False | - |
| 37 | PT08.S2(NMHC) | PT08.S3(NOx) | Granger causality F-test | 21.797713 | 2.606392 | 4.908031e-14 | (3, 5990) | PT08.S2(NMHC) does not Granger-cause PT08.S3(NOx) | reject H_0 at 5% significance level. | True | - |
| 2 | PT08.S3(NOx) | PT08.S3(NOx) | Granger causality F-test | 535.641656 | 2.606392 | 2.171038e-308 | (3, 5990) | PT08.S3(NOx) does not Granger-cause PT08.S3(NOx) | reject H_0 at 5% significance level. | True | - |
| 38 | PT08.S4(NO2) | PT08.S3(NOx) | Granger causality F-test | 1.062097 | 2.606392 | 3.638691e-01 | (3, 5990) | PT08.S4(NO2) does not Granger-cause PT08.S3(NOx) | fail to reject H_0 at 5% significance level. | False | - |
| 39 | PT08.S5(O3) | PT08.S3(NOx) | Granger causality F-test | 4.458826 | 2.606392 | 3.914461e-03 | (3, 5990) | PT08.S5(O3) does not Granger-cause PT08.S3(NOx) | reject H_0 at 5% significance level. | True | - |
| 35 | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\', \'PT08.S4(NO2)\', \'PT08.S5(O3)\'] | PT08.S3(NOx) | Granger causality F-test | 53.823814 | 1.753786 | 1.145878e-123 | (12, 5990) | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\', \'PT08.S4(NO2)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S3(NOx) | reject H_0 at 5% significance level. | True | - |
| 46 | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\', \'PT08.S4(NO2)\'] | PT08.S3(NOx) | Granger causality F-test | 31.062981 | 1.881443 | 1.130043e-53 | (9, 5990) | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\', \'PT08.S4(NO2)\'] do not Granger-cause PT08.S3(NOx) | reject H_0 at 5% significance level. | True | - |
| 47 | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\', \'PT08.S5(O3)\'] | PT08.S3(NOx) | Granger causality F-test | 13.106776 | 1.881443 | 5.800537e-21 | (9, 5990) | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S3(NOx) | reject H_0 at 5% significance level. | True | - |
| 40 | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\'] | PT08.S3(NOx) | Granger causality F-test | 12.417211 | 2.100103 | 5.968478e-14 | (6, 5990) | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\'] do not Granger-cause PT08.S3(NOx) | reject H_0 at 5% significance level. | True | - |
| 48 | [\'PT08.S1(CO)\', \'PT08.S4(NO2)\', \'PT08.S5(O3)\'] | PT08.S3(NOx) | Granger causality F-test | 2.415664 | 1.881443 | 9.829935e-03 | (9, 5990) | [\'PT08.S1(CO)\', \'PT08.S4(NO2)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S3(NOx) | reject H_0 at 5% significance level. | True | - |
| 41 | [\'PT08.S1(CO)\', \'PT08.S4(NO2)\'] | PT08.S3(NOx) | Granger causality F-test | 0.837058 | 2.100103 | 5.410050e-01 | (6, 5990) | [\'PT08.S1(CO)\', \'PT08.S4(NO2)\'] do not Granger-cause PT08.S3(NOx) | fail to reject H_0 at 5% significance level. | False | - |
| 42 | [\'PT08.S1(CO)\', \'PT08.S5(O3)\'] | PT08.S3(NOx) | Granger causality F-test | 3.241024 | 2.100103 | 3.508490e-03 | (6, 5990) | [\'PT08.S1(CO)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S3(NOx) | reject H_0 at 5% significance level. | True | - |
| 49 | [\'PT08.S2(NMHC)\', \'PT08.S4(NO2)\', \'PT08.S5(O3)\'] | PT08.S3(NOx) | Granger causality F-test | 22.849300 | 1.881443 | 1.086013e-38 | (9, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S4(NO2)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S3(NOx) | reject H_0 at 5% significance level. | True | - |
| 43 | [\'PT08.S2(NMHC)\', \'PT08.S4(NO2)\'] | PT08.S3(NOx) | Granger causality F-test | 27.359396 | 2.100103 | 2.231248e-32 | (6, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S4(NO2)\'] do not Granger-cause PT08.S3(NOx) | reject H_0 at 5% significance level. | True | - |
| 44 | [\'PT08.S2(NMHC)\', \'PT08.S5(O3)\'] | PT08.S3(NOx) | Granger causality F-test | 14.794095 | 2.100103 | 7.362809e-17 | (6, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S3(NOx) | reject H_0 at 5% significance level. | True | - |
| 45 | [\'PT08.S4(NO2)\', \'PT08.S5(O3)\'] | PT08.S3(NOx) | Granger causality F-test | 2.831712 | 2.100103 | 9.387131e-03 | (6, 5990) | [\'PT08.S4(NO2)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S3(NOx) | reject H_0 at 5% significance level. | True | - |
| 51 | PT08.S1(CO) | PT08.S4(NO2) | Granger causality F-test | 2.039496 | 2.606392 | 1.061067e-01 | (3, 5990) | PT08.S1(CO) does not Granger-cause PT08.S4(NO2) | fail to reject H_0 at 5% significance level. | False | - |
| 52 | PT08.S2(NMHC) | PT08.S4(NO2) | Granger causality F-test | 52.923585 | 2.606392 | 9.370835e-34 | (3, 5990) | PT08.S2(NMHC) does not Granger-cause PT08.S4(NO2) | reject H_0 at 5% significance level. | True | - |
| 53 | PT08.S3(NOx) | PT08.S4(NO2) | Granger causality F-test | 43.001065 | 2.606392 | 1.740571e-27 | (3, 5990) | PT08.S3(NOx) does not Granger-cause PT08.S4(NO2) | reject H_0 at 5% significance level. | True | - |
| 3 | PT08.S4(NO2) | PT08.S4(NO2) | Granger causality F-test | 30.517211 | 2.606392 | 1.426956e-19 | (3, 5990) | PT08.S4(NO2) does not Granger-cause PT08.S4(NO2) | reject H_0 at 5% significance level. | True | - |
| 54 | PT08.S5(O3) | PT08.S4(NO2) | Granger causality F-test | 0.128394 | 2.606392 | 9.432819e-01 | (3, 5990) | PT08.S5(O3) does not Granger-cause PT08.S4(NO2) | fail to reject H_0 at 5% significance level. | False | - |
| 50 | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\', \'PT08.S3(NOx)\', \'PT08.S5(O3)\'] | PT08.S4(NO2) | Granger causality F-test | 17.302838 | 1.753786 | 4.248405e-37 | (12, 5990) | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\', \'PT08.S3(NOx)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S4(NO2) | reject H_0 at 5% significance level. | True | - |
| 61 | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\', \'PT08.S3(NOx)\'] | PT08.S4(NO2) | Granger causality F-test | 22.805736 | 1.881443 | 1.304286e-38 | (9, 5990) | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\', \'PT08.S3(NOx)\'] do not Granger-cause PT08.S4(NO2) | reject H_0 at 5% significance level. | True | - |
| 62 | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\', \'PT08.S5(O3)\'] | PT08.S4(NO2) | Granger causality F-test | 17.947715 | 1.881443 | 9.511340e-30 | (9, 5990) | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S4(NO2) | reject H_0 at 5% significance level. | True | - |
| 55 | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\'] | PT08.S4(NO2) | Granger causality F-test | 26.550076 | 2.100103 | 2.239768e-31 | (6, 5990) | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\'] do not Granger-cause PT08.S4(NO2) | reject H_0 at 5% significance level. | True | - |
| 63 | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\', \'PT08.S5(O3)\'] | PT08.S4(NO2) | Granger causality F-test | 15.706993 | 1.881443 | 1.133385e-25 | (9, 5990) | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S4(NO2) | reject H_0 at 5% significance level. | True | - |
| 56 | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\'] | PT08.S4(NO2) | Granger causality F-test | 21.614024 | 2.100103 | 2.869846e-25 | (6, 5990) | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\'] do not Granger-cause PT08.S4(NO2) | reject H_0 at 5% significance level. | True | - |
| 57 | [\'PT08.S1(CO)\', \'PT08.S5(O3)\'] | PT08.S4(NO2) | Granger causality F-test | 1.244474 | 2.100103 | 2.800001e-01 | (6, 5990) | [\'PT08.S1(CO)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S4(NO2) | fail to reject H_0 at 5% significance level. | False | - |
| 64 | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\', \'PT08.S5(O3)\'] | PT08.S4(NO2) | Granger causality F-test | 22.987170 | 1.881443 | 6.082898e-39 | (9, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S4(NO2) | reject H_0 at 5% significance level. | True | - |
| 58 | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\'] | PT08.S4(NO2) | Granger causality F-test | 34.079614 | 2.100103 | 1.089561e-40 | (6, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\'] do not Granger-cause PT08.S4(NO2) | reject H_0 at 5% significance level. | True | - |
| 59 | [\'PT08.S2(NMHC)\', \'PT08.S5(O3)\'] | PT08.S4(NO2) | Granger causality F-test | 26.805440 | 2.100103 | 1.081801e-31 | (6, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S4(NO2) | reject H_0 at 5% significance level. | True | - |
| 60 | [\'PT08.S3(NOx)\', \'PT08.S5(O3)\'] | PT08.S4(NO2) | Granger causality F-test | 22.897668 | 2.100103 | 7.415814e-27 | (6, 5990) | [\'PT08.S3(NOx)\', \'PT08.S5(O3)\'] do not Granger-cause PT08.S4(NO2) | reject H_0 at 5% significance level. | True | - |
| 66 | PT08.S1(CO) | PT08.S5(O3) | Granger causality F-test | 0.988534 | 2.606392 | 3.970351e-01 | (3, 5990) | PT08.S1(CO) does not Granger-cause PT08.S5(O3) | fail to reject H_0 at 5% significance level. | False | - |
| 67 | PT08.S2(NMHC) | PT08.S5(O3) | Granger causality F-test | 39.451808 | 2.606392 | 3.076235e-25 | (3, 5990) | PT08.S2(NMHC) does not Granger-cause PT08.S5(O3) | reject H_0 at 5% significance level. | True | - |
| 68 | PT08.S3(NOx) | PT08.S5(O3) | Granger causality F-test | 12.606204 | 2.606392 | 3.266968e-08 | (3, 5990) | PT08.S3(NOx) does not Granger-cause PT08.S5(O3) | reject H_0 at 5% significance level. | True | - |
| 69 | PT08.S4(NO2) | PT08.S5(O3) | Granger causality F-test | 6.255051 | 2.606392 | 3.097984e-04 | (3, 5990) | PT08.S4(NO2) does not Granger-cause PT08.S5(O3) | reject H_0 at 5% significance level. | True | - |
| 4 | PT08.S5(O3) | PT08.S5(O3) | Granger causality F-test | 319.312567 | 2.606392 | 2.487520e-192 | (3, 5990) | PT08.S5(O3) does not Granger-cause PT08.S5(O3) | reject H_0 at 5% significance level. | True | - |
| 65 | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\', \'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] | PT08.S5(O3) | Granger causality F-test | 48.150887 | 1.753786 | 1.790279e-110 | (12, 5990) | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\', \'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] do not Granger-cause PT08.S5(O3) | reject H_0 at 5% significance level. | True | - |
| 76 | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\', \'PT08.S3(NOx)\'] | PT08.S5(O3) | Granger causality F-test | 15.373700 | 1.881443 | 4.565604e-25 | (9, 5990) | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\', \'PT08.S3(NOx)\'] do not Granger-cause PT08.S5(O3) | reject H_0 at 5% significance level. | True | - |
| 77 | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\', \'PT08.S4(NO2)\'] | PT08.S5(O3) | Granger causality F-test | 60.731576 | 1.881443 | 5.676010e-107 | (9, 5990) | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\', \'PT08.S4(NO2)\'] do not Granger-cause PT08.S5(O3) | reject H_0 at 5% significance level. | True | - |
| 70 | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\'] | PT08.S5(O3) | Granger causality F-test | 22.194135 | 2.100103 | 5.500853e-26 | (6, 5990) | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\'] do not Granger-cause PT08.S5(O3) | reject H_0 at 5% significance level. | True | - |
| 78 | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] | PT08.S5(O3) | Granger causality F-test | 7.353858 | 1.881443 | 9.811750e-11 | (9, 5990) | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] do not Granger-cause PT08.S5(O3) | reject H_0 at 5% significance level. | True | - |
| 71 | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\'] | PT08.S5(O3) | Granger causality F-test | 7.055542 | 2.100103 | 1.680113e-07 | (6, 5990) | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\'] do not Granger-cause PT08.S5(O3) | reject H_0 at 5% significance level. | True | - |
| 72 | [\'PT08.S1(CO)\', \'PT08.S4(NO2)\'] | PT08.S5(O3) | Granger causality F-test | 3.285629 | 2.100103 | 3.146561e-03 | (6, 5990) | [\'PT08.S1(CO)\', \'PT08.S4(NO2)\'] do not Granger-cause PT08.S5(O3) | reject H_0 at 5% significance level. | True | - |
| 79 | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] | PT08.S5(O3) | Granger causality F-test | 42.089433 | 1.881443 | 1.150750e-73 | (9, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] do not Granger-cause PT08.S5(O3) | reject H_0 at 5% significance level. | True | - |
| 73 | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\'] | PT08.S5(O3) | Granger causality F-test | 20.895980 | 2.100103 | 2.216207e-24 | (6, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\'] do not Granger-cause PT08.S5(O3) | reject H_0 at 5% significance level. | True | - |
| 74 | [\'PT08.S2(NMHC)\', \'PT08.S4(NO2)\'] | PT08.S5(O3) | Granger causality F-test | 58.630591 | 2.100103 | 8.230165e-71 | (6, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S4(NO2)\'] do not Granger-cause PT08.S5(O3) | reject H_0 at 5% significance level. | True | - |
| 75 | [\'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] | PT08.S5(O3) | Granger causality F-test | 10.167651 | 2.100103 | 3.217170e-11 | (6, 5990) | [\'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] do not Granger-cause PT08.S5(O3) | reject H_0 at 5% significance level. | True | - |
| 180 | PT08.S5(O3) | [PT08.S1(CO), PT08.S2(NMHC), PT08.S3(NOx), PT08.S4(NO2)] | Granger causality F-test | 3.697339 | 1.753786 | 1.389086e-05 | (12, 5990) | PT08.S5(O3) does not Granger-cause [PT08.S1(CO), PT08.S2(NMHC), PT08.S3(NOx), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 181 | PT08.S4(NO2) | [PT08.S1(CO), PT08.S2(NMHC), PT08.S3(NOx), PT08.S5(O3)] | Granger causality F-test | 11.949458 | 1.753786 | 2.566745e-24 | (12, 5990) | PT08.S4(NO2) does not Granger-cause [PT08.S1(CO), PT08.S2(NMHC), PT08.S3(NOx), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 151 | PT08.S4(NO2) | [PT08.S1(CO), PT08.S2(NMHC), PT08.S3(NOx)] | Granger causality F-test | 14.970390 | 1.881443 | 2.461643e-24 | (9, 5990) | PT08.S4(NO2) does not Granger-cause [PT08.S1(CO), PT08.S2(NMHC), PT08.S3(NOx)] | reject H_0 at 5% significance level. | True | - |
| 152 | PT08.S5(O3) | [PT08.S1(CO), PT08.S2(NMHC), PT08.S3(NOx)] | Granger causality F-test | 4.739396 | 1.881443 | 2.630749e-06 | (9, 5990) | PT08.S5(O3) does not Granger-cause [PT08.S1(CO), PT08.S2(NMHC), PT08.S3(NOx)] | reject H_0 at 5% significance level. | True | - |
| 150 | [\'PT08.S4(NO2)\', \'PT08.S5(O3)\'] | [PT08.S1(CO), PT08.S2(NMHC), PT08.S3(NOx)] | Granger causality F-test | 9.811097 | 1.605573 | 1.298403e-27 | (18, 5990) | [\'PT08.S4(NO2)\', \'PT08.S5(O3)\'] do not Granger-cause [PT08.S1(CO), PT08.S2(NMHC), PT08.S3(NOx)] | reject H_0 at 5% significance level. | True | - |
| 182 | PT08.S3(NOx) | [PT08.S1(CO), PT08.S2(NMHC), PT08.S4(NO2), PT08.S5(O3)] | Granger causality F-test | 44.192180 | 1.753786 | 3.351272e-101 | (12, 5990) | PT08.S3(NOx) does not Granger-cause [PT08.S1(CO), PT08.S2(NMHC), PT08.S4(NO2), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 154 | PT08.S3(NOx) | [PT08.S1(CO), PT08.S2(NMHC), PT08.S4(NO2)] | Granger causality F-test | 57.248873 | 1.881443 | 8.396912e-101 | (9, 5990) | PT08.S3(NOx) does not Granger-cause [PT08.S1(CO), PT08.S2(NMHC), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 155 | PT08.S5(O3) | [PT08.S1(CO), PT08.S2(NMHC), PT08.S4(NO2)] | Granger causality F-test | 3.353334 | 1.881443 | 4.182664e-04 | (9, 5990) | PT08.S5(O3) does not Granger-cause [PT08.S1(CO), PT08.S2(NMHC), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 153 | [\'PT08.S3(NOx)\', \'PT08.S5(O3)\'] | [PT08.S1(CO), PT08.S2(NMHC), PT08.S4(NO2)] | Granger causality F-test | 30.916547 | 1.605573 | 1.274714e-101 | (18, 5990) | [\'PT08.S3(NOx)\', \'PT08.S5(O3)\'] do not Granger-cause [PT08.S1(CO), PT08.S2(NMHC), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 157 | PT08.S3(NOx) | [PT08.S1(CO), PT08.S2(NMHC), PT08.S5(O3)] | Granger causality F-test | 56.315310 | 1.881443 | 3.821697e-99 | (9, 5990) | PT08.S3(NOx) does not Granger-cause [PT08.S1(CO), PT08.S2(NMHC), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 158 | PT08.S4(NO2) | [PT08.S1(CO), PT08.S2(NMHC), PT08.S5(O3)] | Granger causality F-test | 13.145421 | 1.881443 | 4.939876e-21 | (9, 5990) | PT08.S4(NO2) does not Granger-cause [PT08.S1(CO), PT08.S2(NMHC), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 156 | [\'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] | [PT08.S1(CO), PT08.S2(NMHC), PT08.S5(O3)] | Granger causality F-test | 39.097140 | 1.605573 | 8.096776e-130 | (18, 5990) | [\'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] do not Granger-cause [PT08.S1(CO), PT08.S2(NMHC), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 81 | PT08.S3(NOx) | [PT08.S1(CO), PT08.S2(NMHC)] | Granger causality F-test | 82.017468 | 2.100103 | 5.250221e-99 | (6, 5990) | PT08.S3(NOx) does not Granger-cause [PT08.S1(CO), PT08.S2(NMHC)] | reject H_0 at 5% significance level. | True | - |
| 82 | PT08.S4(NO2) | [PT08.S1(CO), PT08.S2(NMHC)] | Granger causality F-test | 16.953421 | 2.100103 | 1.628265e-19 | (6, 5990) | PT08.S4(NO2) does not Granger-cause [PT08.S1(CO), PT08.S2(NMHC)] | reject H_0 at 5% significance level. | True | - |
| 83 | PT08.S5(O3) | [PT08.S1(CO), PT08.S2(NMHC)] | Granger causality F-test | 4.760208 | 2.100103 | 7.553233e-05 | (6, 5990) | PT08.S5(O3) does not Granger-cause [PT08.S1(CO), PT08.S2(NMHC)] | reject H_0 at 5% significance level. | True | - |
| 80 | [\'PT08.S3(NOx)\', \'PT08.S4(NO2)\', \'PT08.S5(O3)\'] | [PT08.S1(CO), PT08.S2(NMHC)] | Granger causality F-test | 38.794502 | 1.605573 | 8.773917e-129 | (18, 5990) | [\'PT08.S3(NOx)\', \'PT08.S4(NO2)\', \'PT08.S5(O3)\'] do not Granger-cause [PT08.S1(CO), PT08.S2(NMHC)] | reject H_0 at 5% significance level. | True | - |
| 84 | [\'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] | [PT08.S1(CO), PT08.S2(NMHC)] | Granger causality F-test | 55.255555 | 1.753786 | 5.585312e-127 | (12, 5990) | [\'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] do not Granger-cause [PT08.S1(CO), PT08.S2(NMHC)] | reject H_0 at 5% significance level. | True | - |
| 85 | [\'PT08.S3(NOx)\', \'PT08.S5(O3)\'] | [PT08.S1(CO), PT08.S2(NMHC)] | Granger causality F-test | 44.112366 | 1.753786 | 5.160544e-101 | (12, 5990) | [\'PT08.S3(NOx)\', \'PT08.S5(O3)\'] do not Granger-cause [PT08.S1(CO), PT08.S2(NMHC)] | reject H_0 at 5% significance level. | True | - |
| 86 | [\'PT08.S4(NO2)\', \'PT08.S5(O3)\'] | [PT08.S1(CO), PT08.S2(NMHC)] | Granger causality F-test | 10.744076 | 1.753786 | 1.812578e-21 | (12, 5990) | [\'PT08.S4(NO2)\', \'PT08.S5(O3)\'] do not Granger-cause [PT08.S1(CO), PT08.S2(NMHC)] | reject H_0 at 5% significance level. | True | - |
| 183 | PT08.S2(NMHC) | [PT08.S1(CO), PT08.S3(NOx), PT08.S4(NO2), PT08.S5(O3)] | Granger causality F-test | 20.131969 | 1.753786 | 6.863686e-44 | (12, 5990) | PT08.S2(NMHC) does not Granger-cause [PT08.S1(CO), PT08.S3(NOx), PT08.S4(NO2), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 160 | PT08.S2(NMHC) | [PT08.S1(CO), PT08.S3(NOx), PT08.S4(NO2)] | Granger causality F-test | 24.181136 | 1.881443 | 4.019454e-41 | (9, 5990) | PT08.S2(NMHC) does not Granger-cause [PT08.S1(CO), PT08.S3(NOx), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 161 | PT08.S5(O3) | [PT08.S1(CO), PT08.S3(NOx), PT08.S4(NO2)] | Granger causality F-test | 3.390101 | 1.881443 | 3.675082e-04 | (9, 5990) | PT08.S5(O3) does not Granger-cause [PT08.S1(CO), PT08.S3(NOx), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 159 | [\'PT08.S2(NMHC)\', \'PT08.S5(O3)\'] | [PT08.S1(CO), PT08.S3(NOx), PT08.S4(NO2)] | Granger causality F-test | 13.990940 | 1.605573 | 3.297766e-42 | (18, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S5(O3)\'] do not Granger-cause [PT08.S1(CO), PT08.S3(NOx), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 163 | PT08.S2(NMHC) | [PT08.S1(CO), PT08.S3(NOx), PT08.S5(O3)] | Granger causality F-test | 17.853970 | 1.881443 | 1.409287e-29 | (9, 5990) | PT08.S2(NMHC) does not Granger-cause [PT08.S1(CO), PT08.S3(NOx), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 164 | PT08.S4(NO2) | [PT08.S1(CO), PT08.S3(NOx), PT08.S5(O3)] | Granger causality F-test | 11.652685 | 1.881443 | 2.405236e-18 | (9, 5990) | PT08.S4(NO2) does not Granger-cause [PT08.S1(CO), PT08.S3(NOx), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 162 | [\'PT08.S2(NMHC)\', \'PT08.S4(NO2)\'] | [PT08.S1(CO), PT08.S3(NOx), PT08.S5(O3)] | Granger causality F-test | 43.618536 | 1.605573 | 3.359796e-145 | (18, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S4(NO2)\'] do not Granger-cause [PT08.S1(CO), PT08.S3(NOx), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 88 | PT08.S2(NMHC) | [PT08.S1(CO), PT08.S3(NOx)] | Granger causality F-test | 22.610704 | 2.100103 | 1.679418e-26 | (6, 5990) | PT08.S2(NMHC) does not Granger-cause [PT08.S1(CO), PT08.S3(NOx)] | reject H_0 at 5% significance level. | True | - |
| 89 | PT08.S4(NO2) | [PT08.S1(CO), PT08.S3(NOx)] | Granger causality F-test | 16.301834 | 2.100103 | 1.032657e-18 | (6, 5990) | PT08.S4(NO2) does not Granger-cause [PT08.S1(CO), PT08.S3(NOx)] | reject H_0 at 5% significance level. | True | - |
| 90 | PT08.S5(O3) | [PT08.S1(CO), PT08.S3(NOx)] | Granger causality F-test | 4.800594 | 2.100103 | 6.800829e-05 | (6, 5990) | PT08.S5(O3) does not Granger-cause [PT08.S1(CO), PT08.S3(NOx)] | reject H_0 at 5% significance level. | True | - |
| 87 | [\'PT08.S2(NMHC)\', \'PT08.S4(NO2)\', \'PT08.S5(O3)\'] | [PT08.S1(CO), PT08.S3(NOx)] | Granger causality F-test | 45.020610 | 1.605573 | 6.122018e-150 | (18, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S4(NO2)\', \'PT08.S5(O3)\'] do not Granger-cause [PT08.S1(CO), PT08.S3(NOx)] | reject H_0 at 5% significance level. | True | - |
| 91 | [\'PT08.S2(NMHC)\', \'PT08.S4(NO2)\'] | [PT08.S1(CO), PT08.S3(NOx)] | Granger causality F-test | 62.937316 | 1.753786 | 1.261257e-144 | (12, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S4(NO2)\'] do not Granger-cause [PT08.S1(CO), PT08.S3(NOx)] | reject H_0 at 5% significance level. | True | - |
| 92 | [\'PT08.S2(NMHC)\', \'PT08.S5(O3)\'] | [PT08.S1(CO), PT08.S3(NOx)] | Granger causality F-test | 13.964728 | 1.753786 | 4.132860e-29 | (12, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S5(O3)\'] do not Granger-cause [PT08.S1(CO), PT08.S3(NOx)] | reject H_0 at 5% significance level. | True | - |
| 93 | [\'PT08.S4(NO2)\', \'PT08.S5(O3)\'] | [PT08.S1(CO), PT08.S3(NOx)] | Granger causality F-test | 10.548064 | 1.753786 | 5.246873e-21 | (12, 5990) | [\'PT08.S4(NO2)\', \'PT08.S5(O3)\'] do not Granger-cause [PT08.S1(CO), PT08.S3(NOx)] | reject H_0 at 5% significance level. | True | - |
| 166 | PT08.S2(NMHC) | [PT08.S1(CO), PT08.S4(NO2), PT08.S5(O3)] | Granger causality F-test | 26.737321 | 1.881443 | 8.676974e-46 | (9, 5990) | PT08.S2(NMHC) does not Granger-cause [PT08.S1(CO), PT08.S4(NO2), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 167 | PT08.S3(NOx) | [PT08.S1(CO), PT08.S4(NO2), PT08.S5(O3)] | Granger causality F-test | 18.682608 | 1.881443 | 4.356107e-31 | (9, 5990) | PT08.S3(NOx) does not Granger-cause [PT08.S1(CO), PT08.S4(NO2), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 165 | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\'] | [PT08.S1(CO), PT08.S4(NO2), PT08.S5(O3)] | Granger causality F-test | 18.360429 | 1.605573 | 1.332973e-57 | (18, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\'] do not Granger-cause [PT08.S1(CO), PT08.S4(NO2), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 95 | PT08.S2(NMHC) | [PT08.S1(CO), PT08.S4(NO2)] | Granger causality F-test | 35.097800 | 2.100103 | 6.019555e-42 | (6, 5990) | PT08.S2(NMHC) does not Granger-cause [PT08.S1(CO), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 96 | PT08.S3(NOx) | [PT08.S1(CO), PT08.S4(NO2)] | Granger causality F-test | 24.678203 | 2.100103 | 4.644502e-29 | (6, 5990) | PT08.S3(NOx) does not Granger-cause [PT08.S1(CO), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 97 | PT08.S5(O3) | [PT08.S1(CO), PT08.S4(NO2)] | Granger causality F-test | 1.318931 | 2.100103 | 2.446962e-01 | (6, 5990) | PT08.S5(O3) does not Granger-cause [PT08.S1(CO), PT08.S4(NO2)] | fail to reject H_0 at 5% significance level. | False | - |
| 94 | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\', \'PT08.S5(O3)\'] | [PT08.S1(CO), PT08.S4(NO2)] | Granger causality F-test | 16.618862 | 1.605573 | 1.842573e-51 | (18, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\', \'PT08.S5(O3)\'] do not Granger-cause [PT08.S1(CO), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 98 | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\'] | [PT08.S1(CO), PT08.S4(NO2)] | Granger causality F-test | 24.198090 | 1.753786 | 1.177786e-53 | (12, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\'] do not Granger-cause [PT08.S1(CO), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 99 | [\'PT08.S2(NMHC)\', \'PT08.S5(O3)\'] | [PT08.S1(CO), PT08.S4(NO2)] | Granger causality F-test | 18.035714 | 1.753786 | 7.412041e-39 | (12, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S5(O3)\'] do not Granger-cause [PT08.S1(CO), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 100 | [\'PT08.S3(NOx)\', \'PT08.S5(O3)\'] | [PT08.S1(CO), PT08.S4(NO2)] | Granger causality F-test | 14.041399 | 1.753786 | 2.712284e-29 | (12, 5990) | [\'PT08.S3(NOx)\', \'PT08.S5(O3)\'] do not Granger-cause [PT08.S1(CO), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 102 | PT08.S2(NMHC) | [PT08.S1(CO), PT08.S5(O3)] | Granger causality F-test | 25.512310 | 2.100103 | 4.311451e-30 | (6, 5990) | PT08.S2(NMHC) does not Granger-cause [PT08.S1(CO), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 103 | PT08.S3(NOx) | [PT08.S1(CO), PT08.S5(O3)] | Granger causality F-test | 10.227499 | 2.100103 | 2.723753e-11 | (6, 5990) | PT08.S3(NOx) does not Granger-cause [PT08.S1(CO), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 104 | PT08.S4(NO2) | [PT08.S1(CO), PT08.S5(O3)] | Granger causality F-test | 16.801326 | 2.100103 | 2.506390e-19 | (6, 5990) | PT08.S4(NO2) does not Granger-cause [PT08.S1(CO), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 101 | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] | [PT08.S1(CO), PT08.S5(O3)] | Granger causality F-test | 49.970543 | 1.605573 | 1.485641e-166 | (18, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] do not Granger-cause [PT08.S1(CO), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 105 | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\'] | [PT08.S1(CO), PT08.S5(O3)] | Granger causality F-test | 15.694140 | 1.753786 | 3.041488e-33 | (12, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\'] do not Granger-cause [PT08.S1(CO), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 106 | [\'PT08.S2(NMHC)\', \'PT08.S4(NO2)\'] | [PT08.S1(CO), PT08.S5(O3)] | Granger causality F-test | 64.790015 | 1.753786 | 7.522899e-149 | (12, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S4(NO2)\'] do not Granger-cause [PT08.S1(CO), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 107 | [\'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] | [PT08.S1(CO), PT08.S5(O3)] | Granger causality F-test | 14.501687 | 1.753786 | 2.160320e-30 | (12, 5990) | [\'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] do not Granger-cause [PT08.S1(CO), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 184 | PT08.S1(CO) | [PT08.S2(NMHC), PT08.S3(NOx), PT08.S4(NO2), PT08.S5(O3)] | Granger causality F-test | 2.206276 | 1.753786 | 9.298224e-03 | (12, 5990) | PT08.S1(CO) does not Granger-cause [PT08.S2(NMHC), PT08.S3(NOx), PT08.S4(NO2), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 169 | PT08.S1(CO) | [PT08.S2(NMHC), PT08.S3(NOx), PT08.S4(NO2)] | Granger causality F-test | 2.662247 | 1.881443 | 4.419203e-03 | (9, 5990) | PT08.S1(CO) does not Granger-cause [PT08.S2(NMHC), PT08.S3(NOx), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 170 | PT08.S5(O3) | [PT08.S2(NMHC), PT08.S3(NOx), PT08.S4(NO2)] | Granger causality F-test | 3.007388 | 1.881443 | 1.387469e-03 | (9, 5990) | PT08.S5(O3) does not Granger-cause [PT08.S2(NMHC), PT08.S3(NOx), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 168 | [\'PT08.S1(CO)\', \'PT08.S5(O3)\'] | [PT08.S2(NMHC), PT08.S3(NOx), PT08.S4(NO2)] | Granger causality F-test | 2.935470 | 1.605573 | 2.943222e-05 | (18, 5990) | [\'PT08.S1(CO)\', \'PT08.S5(O3)\'] do not Granger-cause [PT08.S2(NMHC), PT08.S3(NOx), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 172 | PT08.S1(CO) | [PT08.S2(NMHC), PT08.S3(NOx), PT08.S5(O3)] | Granger causality F-test | 2.294625 | 1.881443 | 1.440787e-02 | (9, 5990) | PT08.S1(CO) does not Granger-cause [PT08.S2(NMHC), PT08.S3(NOx), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 173 | PT08.S4(NO2) | [PT08.S2(NMHC), PT08.S3(NOx), PT08.S5(O3)] | Granger causality F-test | 8.009162 | 1.881443 | 7.080145e-12 | (9, 5990) | PT08.S4(NO2) does not Granger-cause [PT08.S2(NMHC), PT08.S3(NOx), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 171 | [\'PT08.S1(CO)\', \'PT08.S4(NO2)\'] | [PT08.S2(NMHC), PT08.S3(NOx), PT08.S5(O3)] | Granger causality F-test | 4.806738 | 1.605573 | 7.435402e-11 | (18, 5990) | [\'PT08.S1(CO)\', \'PT08.S4(NO2)\'] do not Granger-cause [PT08.S2(NMHC), PT08.S3(NOx), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 109 | PT08.S1(CO) | [PT08.S2(NMHC), PT08.S3(NOx)] | Granger causality F-test | 3.068935 | 2.100103 | 5.324794e-03 | (6, 5990) | PT08.S1(CO) does not Granger-cause [PT08.S2(NMHC), PT08.S3(NOx)] | reject H_0 at 5% significance level. | True | - |
| 110 | PT08.S4(NO2) | [PT08.S2(NMHC), PT08.S3(NOx)] | Granger causality F-test | 9.091622 | 2.100103 | 6.355526e-10 | (6, 5990) | PT08.S4(NO2) does not Granger-cause [PT08.S2(NMHC), PT08.S3(NOx)] | reject H_0 at 5% significance level. | True | - |
| 111 | PT08.S5(O3) | [PT08.S2(NMHC), PT08.S3(NOx)] | Granger causality F-test | 4.011977 | 2.100103 | 5.153055e-04 | (6, 5990) | PT08.S5(O3) does not Granger-cause [PT08.S2(NMHC), PT08.S3(NOx)] | reject H_0 at 5% significance level. | True | - |
| 108 | [\'PT08.S1(CO)\', \'PT08.S4(NO2)\', \'PT08.S5(O3)\'] | [PT08.S2(NMHC), PT08.S3(NOx)] | Granger causality F-test | 5.142349 | 1.605573 | 6.366177e-12 | (18, 5990) | [\'PT08.S1(CO)\', \'PT08.S4(NO2)\', \'PT08.S5(O3)\'] do not Granger-cause [PT08.S2(NMHC), PT08.S3(NOx)] | reject H_0 at 5% significance level. | True | - |
| 112 | [\'PT08.S1(CO)\', \'PT08.S4(NO2)\'] | [PT08.S2(NMHC), PT08.S3(NOx)] | Granger causality F-test | 5.697091 | 1.753786 | 7.455053e-10 | (12, 5990) | [\'PT08.S1(CO)\', \'PT08.S4(NO2)\'] do not Granger-cause [PT08.S2(NMHC), PT08.S3(NOx)] | reject H_0 at 5% significance level. | True | - |
| 113 | [\'PT08.S1(CO)\', \'PT08.S5(O3)\'] | [PT08.S2(NMHC), PT08.S3(NOx)] | Granger causality F-test | 3.252565 | 1.753786 | 1.077941e-04 | (12, 5990) | [\'PT08.S1(CO)\', \'PT08.S5(O3)\'] do not Granger-cause [PT08.S2(NMHC), PT08.S3(NOx)] | reject H_0 at 5% significance level. | True | - |
| 114 | [\'PT08.S4(NO2)\', \'PT08.S5(O3)\'] | [PT08.S2(NMHC), PT08.S3(NOx)] | Granger causality F-test | 6.711379 | 1.753786 | 4.008284e-12 | (12, 5990) | [\'PT08.S4(NO2)\', \'PT08.S5(O3)\'] do not Granger-cause [PT08.S2(NMHC), PT08.S3(NOx)] | reject H_0 at 5% significance level. | True | - |
| 175 | PT08.S1(CO) | [PT08.S2(NMHC), PT08.S4(NO2), PT08.S5(O3)] | Granger causality F-test | 1.395341 | 1.881443 | 1.839347e-01 | (9, 5990) | PT08.S1(CO) does not Granger-cause [PT08.S2(NMHC), PT08.S4(NO2), PT08.S5(O3)] | fail to reject H_0 at 5% significance level. | False | - |
| 176 | PT08.S3(NOx) | [PT08.S2(NMHC), PT08.S4(NO2), PT08.S5(O3)] | Granger causality F-test | 52.342075 | 1.881443 | 4.559191e-92 | (9, 5990) | PT08.S3(NOx) does not Granger-cause [PT08.S2(NMHC), PT08.S4(NO2), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 174 | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\'] | [PT08.S2(NMHC), PT08.S4(NO2), PT08.S5(O3)] | Granger causality F-test | 26.954125 | 1.605573 | 8.274599e-88 | (18, 5990) | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\'] do not Granger-cause [PT08.S2(NMHC), PT08.S4(NO2), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 116 | PT08.S1(CO) | [PT08.S2(NMHC), PT08.S4(NO2)] | Granger causality F-test | 1.538218 | 2.100103 | 1.612991e-01 | (6, 5990) | PT08.S1(CO) does not Granger-cause [PT08.S2(NMHC), PT08.S4(NO2)] | fail to reject H_0 at 5% significance level. | False | - |
| 117 | PT08.S3(NOx) | [PT08.S2(NMHC), PT08.S4(NO2)] | Granger causality F-test | 73.385337 | 2.100103 | 1.162926e-88 | (6, 5990) | PT08.S3(NOx) does not Granger-cause [PT08.S2(NMHC), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 118 | PT08.S5(O3) | [PT08.S2(NMHC), PT08.S4(NO2)] | Granger causality F-test | 2.064536 | 2.100103 | 5.403727e-02 | (6, 5990) | PT08.S5(O3) does not Granger-cause [PT08.S2(NMHC), PT08.S4(NO2)] | fail to reject H_0 at 5% significance level. | False | - |
| 115 | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\', \'PT08.S5(O3)\'] | [PT08.S2(NMHC), PT08.S4(NO2)] | Granger causality F-test | 26.836549 | 1.605573 | 2.132832e-87 | (18, 5990) | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\', \'PT08.S5(O3)\'] do not Granger-cause [PT08.S2(NMHC), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 119 | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\'] | [PT08.S2(NMHC), PT08.S4(NO2)] | Granger causality F-test | 37.753857 | 1.753786 | 5.186991e-86 | (12, 5990) | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\'] do not Granger-cause [PT08.S2(NMHC), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 120 | [\'PT08.S1(CO)\', \'PT08.S5(O3)\'] | [PT08.S2(NMHC), PT08.S4(NO2)] | Granger causality F-test | 2.616769 | 1.753786 | 1.744625e-03 | (12, 5990) | [\'PT08.S1(CO)\', \'PT08.S5(O3)\'] do not Granger-cause [PT08.S2(NMHC), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 121 | [\'PT08.S3(NOx)\', \'PT08.S5(O3)\'] | [PT08.S2(NMHC), PT08.S4(NO2)] | Granger causality F-test | 38.312533 | 1.753786 | 2.464349e-87 | (12, 5990) | [\'PT08.S3(NOx)\', \'PT08.S5(O3)\'] do not Granger-cause [PT08.S2(NMHC), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 123 | PT08.S1(CO) | [PT08.S2(NMHC), PT08.S5(O3)] | Granger causality F-test | 1.143730 | 2.100103 | 3.339180e-01 | (6, 5990) | PT08.S1(CO) does not Granger-cause [PT08.S2(NMHC), PT08.S5(O3)] | fail to reject H_0 at 5% significance level. | False | - |
| 124 | PT08.S3(NOx) | [PT08.S2(NMHC), PT08.S5(O3)] | Granger causality F-test | 75.963406 | 2.100103 | 9.292988e-92 | (6, 5990) | PT08.S3(NOx) does not Granger-cause [PT08.S2(NMHC), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 125 | PT08.S4(NO2) | [PT08.S2(NMHC), PT08.S5(O3)] | Granger causality F-test | 7.066363 | 2.100103 | 1.631540e-07 | (6, 5990) | PT08.S4(NO2) does not Granger-cause [PT08.S2(NMHC), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 122 | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] | [PT08.S2(NMHC), PT08.S5(O3)] | Granger causality F-test | 35.354601 | 1.605573 | 5.643814e-117 | (18, 5990) | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] do not Granger-cause [PT08.S2(NMHC), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 126 | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\'] | [PT08.S2(NMHC), PT08.S5(O3)] | Granger causality F-test | 38.886015 | 1.753786 | 1.082449e-88 | (12, 5990) | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\'] do not Granger-cause [PT08.S2(NMHC), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 127 | [\'PT08.S1(CO)\', \'PT08.S4(NO2)\'] | [PT08.S2(NMHC), PT08.S5(O3)] | Granger causality F-test | 3.869675 | 1.753786 | 6.170090e-06 | (12, 5990) | [\'PT08.S1(CO)\', \'PT08.S4(NO2)\'] do not Granger-cause [PT08.S2(NMHC), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 128 | [\'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] | [PT08.S2(NMHC), PT08.S5(O3)] | Granger causality F-test | 49.798629 | 1.753786 | 2.565633e-114 | (12, 5990) | [\'PT08.S3(NOx)\', \'PT08.S4(NO2)\'] do not Granger-cause [PT08.S2(NMHC), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 178 | PT08.S1(CO) | [PT08.S3(NOx), PT08.S4(NO2), PT08.S5(O3)] | Granger causality F-test | 1.501865 | 1.881443 | 1.408765e-01 | (9, 5990) | PT08.S1(CO) does not Granger-cause [PT08.S3(NOx), PT08.S4(NO2), PT08.S5(O3)] | fail to reject H_0 at 5% significance level. | False | - |
| 179 | PT08.S2(NMHC) | [PT08.S3(NOx), PT08.S4(NO2), PT08.S5(O3)] | Granger causality F-test | 25.315687 | 1.881443 | 3.411476e-43 | (9, 5990) | PT08.S2(NMHC) does not Granger-cause [PT08.S3(NOx), PT08.S4(NO2), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 177 | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\'] | [PT08.S3(NOx), PT08.S4(NO2), PT08.S5(O3)] | Granger causality F-test | 13.804226 | 1.605573 | 1.493988e-41 | (18, 5990) | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\'] do not Granger-cause [PT08.S3(NOx), PT08.S4(NO2), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 130 | PT08.S1(CO) | [PT08.S3(NOx), PT08.S4(NO2)] | Granger causality F-test | 1.768309 | 2.100103 | 1.014136e-01 | (6, 5990) | PT08.S1(CO) does not Granger-cause [PT08.S3(NOx), PT08.S4(NO2)] | fail to reject H_0 at 5% significance level. | False | - |
| 131 | PT08.S2(NMHC) | [PT08.S3(NOx), PT08.S4(NO2)] | Granger causality F-test | 29.753911 | 2.100103 | 2.429565e-35 | (6, 5990) | PT08.S2(NMHC) does not Granger-cause [PT08.S3(NOx), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 132 | PT08.S5(O3) | [PT08.S3(NOx), PT08.S4(NO2)] | Granger causality F-test | 2.734038 | 2.100103 | 1.181956e-02 | (6, 5990) | PT08.S5(O3) does not Granger-cause [PT08.S3(NOx), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 129 | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\', \'PT08.S5(O3)\'] | [PT08.S3(NOx), PT08.S4(NO2)] | Granger causality F-test | 13.045242 | 1.605573 | 6.898592e-39 | (18, 5990) | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\', \'PT08.S5(O3)\'] do not Granger-cause [PT08.S3(NOx), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 133 | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\'] | [PT08.S3(NOx), PT08.S4(NO2)] | Granger causality F-test | 15.745941 | 1.753786 | 2.286051e-33 | (12, 5990) | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\'] do not Granger-cause [PT08.S3(NOx), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 134 | [\'PT08.S1(CO)\', \'PT08.S5(O3)\'] | [PT08.S3(NOx), PT08.S4(NO2)] | Granger causality F-test | 2.853525 | 1.753786 | 6.333739e-04 | (12, 5990) | [\'PT08.S1(CO)\', \'PT08.S5(O3)\'] do not Granger-cause [PT08.S3(NOx), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 135 | [\'PT08.S2(NMHC)\', \'PT08.S5(O3)\'] | [PT08.S3(NOx), PT08.S4(NO2)] | Granger causality F-test | 16.988859 | 1.753786 | 2.405278e-36 | (12, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S5(O3)\'] do not Granger-cause [PT08.S3(NOx), PT08.S4(NO2)] | reject H_0 at 5% significance level. | True | - |
| 137 | PT08.S1(CO) | [PT08.S3(NOx), PT08.S5(O3)] | Granger causality F-test | 0.758355 | 2.100103 | 6.026898e-01 | (6, 5990) | PT08.S1(CO) does not Granger-cause [PT08.S3(NOx), PT08.S5(O3)] | fail to reject H_0 at 5% significance level. | False | - |
| 138 | PT08.S2(NMHC) | [PT08.S3(NOx), PT08.S5(O3)] | Granger causality F-test | 21.837842 | 2.100103 | 1.517333e-25 | (6, 5990) | PT08.S2(NMHC) does not Granger-cause [PT08.S3(NOx), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 139 | PT08.S4(NO2) | [PT08.S3(NOx), PT08.S5(O3)] | Granger causality F-test | 3.183756 | 2.100103 | 4.033080e-03 | (6, 5990) | PT08.S4(NO2) does not Granger-cause [PT08.S3(NOx), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 136 | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\', \'PT08.S4(NO2)\'] | [PT08.S3(NOx), PT08.S5(O3)] | Granger causality F-test | 32.967278 | 1.605573 | 9.876680e-109 | (18, 5990) | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\', \'PT08.S4(NO2)\'] do not Granger-cause [PT08.S3(NOx), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 140 | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\'] | [PT08.S3(NOx), PT08.S5(O3)] | Granger causality F-test | 12.465452 | 1.753786 | 1.531878e-25 | (12, 5990) | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\'] do not Granger-cause [PT08.S3(NOx), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 141 | [\'PT08.S1(CO)\', \'PT08.S4(NO2)\'] | [PT08.S3(NOx), PT08.S5(O3)] | Granger causality F-test | 1.786970 | 1.753786 | 4.451629e-02 | (12, 5990) | [\'PT08.S1(CO)\', \'PT08.S4(NO2)\'] do not Granger-cause [PT08.S3(NOx), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 142 | [\'PT08.S2(NMHC)\', \'PT08.S4(NO2)\'] | [PT08.S3(NOx), PT08.S5(O3)] | Granger causality F-test | 31.224285 | 1.753786 | 1.763191e-70 | (12, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S4(NO2)\'] do not Granger-cause [PT08.S3(NOx), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 144 | PT08.S1(CO) | [PT08.S4(NO2), PT08.S5(O3)] | Granger causality F-test | 1.667514 | 2.100103 | 1.246490e-01 | (6, 5990) | PT08.S1(CO) does not Granger-cause [PT08.S4(NO2), PT08.S5(O3)] | fail to reject H_0 at 5% significance level. | False | - |
| 145 | PT08.S2(NMHC) | [PT08.S4(NO2), PT08.S5(O3)] | Granger causality F-test | 37.730640 | 2.100103 | 3.388744e-45 | (6, 5990) | PT08.S2(NMHC) does not Granger-cause [PT08.S4(NO2), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 146 | PT08.S3(NOx) | [PT08.S4(NO2), PT08.S5(O3)] | Granger causality F-test | 24.046453 | 2.100103 | 2.810268e-28 | (6, 5990) | PT08.S3(NOx) does not Granger-cause [PT08.S4(NO2), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 143 | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\', \'PT08.S3(NOx)\'] | [PT08.S4(NO2), PT08.S5(O3)] | Granger causality F-test | 16.449647 | 1.605573 | 7.276024e-51 | (18, 5990) | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\', \'PT08.S3(NOx)\'] do not Granger-cause [PT08.S4(NO2), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 147 | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\'] | [PT08.S4(NO2), PT08.S5(O3)] | Granger causality F-test | 20.214069 | 1.753786 | 4.358684e-44 | (12, 5990) | [\'PT08.S1(CO)\', \'PT08.S2(NMHC)\'] do not Granger-cause [PT08.S4(NO2), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 148 | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\'] | [PT08.S4(NO2), PT08.S5(O3)] | Granger causality F-test | 12.463419 | 1.753786 | 1.548994e-25 | (12, 5990) | [\'PT08.S1(CO)\', \'PT08.S3(NOx)\'] do not Granger-cause [PT08.S4(NO2), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 149 | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\'] | [PT08.S4(NO2), PT08.S5(O3)] | Granger causality F-test | 23.473280 | 1.753786 | 6.473219e-52 | (12, 5990) | [\'PT08.S2(NMHC)\', \'PT08.S3(NOx)\'] do not Granger-cause [PT08.S4(NO2), PT08.S5(O3)] | reject H_0 at 5% significance level. | True | - |
| 189 | PT08.S5(O3) | [PT08.S1(CO), PT08.S2(NMHC), PT08.S3(NOx), PT08.S4(NO2)] | Instantaneous causality Wald-test | 337.439976 | 9.487729 | 9.027617e-72 | 4 | PT08.S5(O3) does not instantaneously cause [PT08.S1(CO), PT08.S2(NMHC), PT08.S3(NOx), PT08.S4(NO2)] | reject H_0 at 5% significance level. | - | True |
| 188 | PT08.S4(NO2) | [PT08.S1(CO), PT08.S2(NMHC), PT08.S3(NOx), PT08.S5(O3)] | Instantaneous causality Wald-test | 217.970810 | 9.487729 | 5.123598e-46 | 4 | PT08.S4(NO2) does not instantaneously cause [PT08.S1(CO), PT08.S2(NMHC), PT08.S3(NOx), PT08.S5(O3)] | reject H_0 at 5% significance level. | - | True |
| 187 | PT08.S3(NOx) | [PT08.S1(CO), PT08.S2(NMHC), PT08.S4(NO2), PT08.S5(O3)] | Instantaneous causality Wald-test | 460.331063 | 9.487729 | 2.536898e-98 | 4 | PT08.S3(NOx) does not instantaneously cause [PT08.S1(CO), PT08.S2(NMHC), PT08.S4(NO2), PT08.S5(O3)] | reject H_0 at 5% significance level. | - | True |
| 186 | PT08.S2(NMHC) | [PT08.S1(CO), PT08.S3(NOx), PT08.S4(NO2), PT08.S5(O3)] | Instantaneous causality Wald-test | 486.285247 | 9.487729 | 6.196463e-104 | 4 | PT08.S2(NMHC) does not instantaneously cause [PT08.S1(CO), PT08.S3(NOx), PT08.S4(NO2), PT08.S5(O3)] | reject H_0 at 5% significance level. | - | True |
| 185 | PT08.S1(CO) | [PT08.S2(NMHC), PT08.S3(NOx), PT08.S4(NO2), PT08.S5(O3)] | Instantaneous causality Wald-test | 351.004087 | 9.487729 | 1.064586e-74 | 4 | PT08.S1(CO) does not instantaneously cause [PT08.S2(NMHC), PT08.S3(NOx), PT08.S4(NO2), PT08.S5(O3)] | reject H_0 at 5% significance level. | - | True |
Time Series Model Evaluation
Baseline Explanation Model

- Naive Forecast, ETS(AAN)
- Target(All, 13): 'PT08.S1(CO)', 'PT08.S2(NMHC)', 'PT08.S3(NOx)', 'PT08.S4(NO2)', 'PT08.S5(O3)', 'CO(GT)', 'NMHC(GT)', 'C6H6(GT)', 'NOx(GT)', 'NO2(GT)', 'T', 'RH', 'AH'
- h-step forecast evaluation
- [Train Period]: 2004-03-10 18:00:00 ~ 2004-04-27 23:00:00, 1158
- [Test Period]: 2004-04-28 00:00:00 ~ 2004-05-01 00:00:00, 73
| Model | Naive | ETS AAdN | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Domain | Train | Test | Train | Test | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Info | start | end | model | total | sst | ssr | sse | ssr/sst | sse/sst | ssr/sse | mst | msr | mse | rmse | mape | start | end | model | total | sst | ssr | sse | ssr/sst | sse/sst | ssr/sse | mst | msr | mse | rmse | mape | start | end | total | sst | ssr | sse | ssr/sst | sse/sst | ssr/sse | mst | msr | mse | rmse | mape | start | end | total | sst | ssr | sse | ssr/sst | sse/sst | ssr/sse | mst | msr | mse | rmse | mape |
| CO(GT) | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Naive | 1158 | 952.01381 | 952.551014 | 472.575238 | 1.000564 | 0.496395 | 2.01566 | 0.82283 | 952.551014 | 0.408448 | 0.638824 | 17.320883 | 2004-04-28 | 2004-05-01 | Naive | 73 | 952.01381 | 3.052784 | 80.186239 | 0.003207 | 0.084228 | 0.038071 | 0.82283 | 3.052784 | 0.856304 | 1.048066 | 2.18195 | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | 1158 | 952.01381 | 952.954703 | 472.597244 | 1.000988 | 0.496418 | 2.01642 | 0.82283 | 952.954703 | 0.408467 | 0.638839 | 17.321287 | 2004-04-28 | 2004-05-01 | 73 | 61.653863 | 18.527789 | 80.181143 | 0.300513 | 1.300505 | 0.231074 | 0.856304 | 18.527789 | 0.856297 | 1.048032 | 2.182516 |
| PT08.S1(CO) | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Naive | 1158 | 37329666.834193 | 37329601.532309 | 12421823.909175 | 0.999998 | 0.33276 | 3.005163 | 32264.189139 | 37329601.532309 | 10736.234861 | 103.571056 | 2.06029 | 2004-04-28 | 2004-05-01 | Naive | 73 | 37329666.834193 | 6469.483204 | 2978327.785717 | 0.000173 | 0.079784 | 0.002172 | 32264.189139 | 6469.483204 | 18938.320225 | 201.98765 | 2.004873 | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | 1158 | 37329666.834193 | 37336579.645433 | 12422778.357097 | 1.000185 | 0.332786 | 3.005494 | 32264.189139 | 37336579.645433 | 10737.057413 | 103.575035 | 2.065533 | 2004-04-28 | 2004-05-01 | 73 | 1363559.056179 | 1614370.899647 | 2977898.411039 | 1.183939 | 2.183916 | 0.542118 | 18938.320225 | 1614370.899647 | 18937.882121 | 201.97309 | 2.003703 |
| NMHC(GT) | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Naive | 1158 | 21597542.259443 | 21643014.400826 | 10580453.19625 | 1.002105 | 0.489892 | 2.045566 | 18666.847242 | 21643014.400826 | 9144.662762 | 95.586785 | 3.521068 | 2004-04-28 | 2004-05-01 | Naive | 73 | 21597542.259443 | 412474.657532 | 1960016.119058 | 0.019098 | 0.090752 | 0.210445 | 18666.847242 | 412474.657532 | 25530.259559 | 163.85828 | 4.493315 | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | 1158 | 21597542.259443 | 21620296.990207 | 10583569.292906 | 1.001054 | 0.490036 | 2.042817 | 18666.847242 | 21620296.990207 | 9147.397913 | 95.60086 | 3.520907 | 2004-04-28 | 2004-05-01 | 73 | 1838178.688282 | 121729.02287 | 1959896.178863 | 0.066223 | 1.066216 | 0.06211 | 25530.259559 | 121729.02287 | 25530.099417 | 163.853266 | 4.494018 |
| C6H6(GT) | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Naive | 1158 | 32446.767948 | 32454.011441 | 13750.187331 | 1.000223 | 0.423777 | 2.36026 | 28.043879 | 32454.011441 | 11.884298 | 3.445879 | 4.924125 | 2004-04-28 | 2004-05-01 | Naive | 73 | 32446.767948 | 910.434266 | 1852.531235 | 0.028059 | 0.057094 | 0.491454 | 28.043879 | 910.434266 | 22.320845 | 5.037573 | 2.563734 | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | 1158 | 32446.767948 | 32456.543488 | 13750.944468 | 1.000301 | 0.4238 | 2.360314 | 28.043879 | 32456.543488 | 11.884951 | 3.445974 | 4.924325 | 2004-04-28 | 2004-05-01 | 73 | 1607.100815 | 245.334122 | 1852.418523 | 0.152656 | 1.152646 | 0.13244 | 22.320845 | 245.334122 | 22.320617 | 5.03742 | 2.563929 |
| PT08.S2(NMHC) | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Naive | 1158 | 39472603.176647 | 39463600.686054 | 13978758.389615 | 0.999772 | 0.354138 | 2.823112 | 34116.338096 | 39463600.686054 | 12081.845616 | 109.870226 | 3.318938 | 2004-04-28 | 2004-05-01 | Naive | 73 | 39472603.176647 | 1680521.102278 | 1469868.10471 | 0.042574 | 0.037238 | 1.143314 | 34116.338096 | 1680521.102278 | 18994.399669 | 141.898483 | 2.954736 | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | 1158 | 39472603.176647 | 39472289.139354 | 13984909.452211 | 0.999992 | 0.354294 | 2.822492 | 34116.338096 | 39472289.139354 | 12087.154238 | 109.894397 | 3.319558 | 2004-04-28 | 2004-05-01 | 73 | 1367596.776148 | 102180.939834 | 1469753.648307 | 0.074716 | 1.074698 | 0.069522 | 18994.399669 | 102180.939834 | 18994.065414 | 141.892958 | 2.954923 |
| NOx(GT) | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Naive | 1158 | 3560007.78425 | 3560228.362376 | 1590908.257933 | 1.000062 | 0.446883 | 2.237859 | 3076.929805 | 3560228.362376 | 1375.028492 | 37.065366 | 3.012007 | 2004-04-28 | 2004-05-01 | Naive | 73 | 3560007.78425 | 756.619486 | 238699.479019 | 0.000213 | 0.06705 | 0.00317 | 3076.929805 | 756.619486 | 2405.954339 | 57.182654 | 0.988974 | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | 1158 | 3560007.78425 | 3536970.879439 | 1592098.63543 | 0.993529 | 0.447218 | 2.221578 | 3076.929805 | 3536970.879439 | 1376.056657 | 37.07923 | 3.003566 | 2004-04-28 | 2004-05-01 | 73 | 173228.712409 | 67124.454007 | 240351.129963 | 0.38749 | 1.387479 | 0.279277 | 2405.954339 | 67124.454007 | 2405.926056 | 57.380147 | 0.992285 |
| PT08.S3(NOx) | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Naive | 1158 | 42892614.422674 | 42916057.572883 | 11252537.815437 | 1.000547 | 0.262342 | 3.8139 | 37072.2683 | 42916057.572883 | 9725.476366 | 98.575948 | 6.180128 | 2004-04-28 | 2004-05-01 | Naive | 73 | 42892614.422674 | 2614336.554891 | 724128.988102 | 0.060951 | 0.016882 | 3.610319 | 37072.2683 | 2614336.554891 | 8528.843129 | 99.597064 | 0.483034 | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | 1158 | 42892614.422674 | 42958140.960404 | 11253185.294241 | 1.001528 | 0.262357 | 3.817421 | 37072.2683 | 42958140.960404 | 9725.988093 | 98.578784 | 6.180603 | 2004-04-28 | 2004-05-01 | 73 | 614076.705292 | 109857.568531 | 723910.81941 | 0.178899 | 1.178861 | 0.151756 | 8528.843129 | 109857.568531 | 8528.517426 | 99.582059 | 0.483035 |
| NO2(GT) | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Naive | 1158 | 512557.628272 | 512481.325455 | 202092.401412 | 0.999851 | 0.394282 | 2.535876 | 443.005729 | 512481.325455 | 174.668826 | 13.210544 | 3.282934 | 2004-04-28 | 2004-05-01 | Naive | 73 | 512557.628272 | 14975.63331 | 18032.584565 | 0.029217 | 0.035182 | 0.830476 | 443.005729 | 14975.63331 | 219.010147 | 15.716924 | 8.176079 | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | 1158 | 512557.628272 | 491611.511613 | 200763.614401 | 0.959134 | 0.39169 | 2.448708 | 443.005729 | 491611.511613 | 173.520837 | 13.167042 | 3.414677 | 2004-04-28 | 2004-05-01 | 73 | 15768.730578 | 1209.220989 | 16977.628126 | 0.076685 | 1.076664 | 0.071224 | 219.010147 | 1209.220989 | 219.005692 | 15.250254 | 7.337192 |
| PT08.S4(NO2) | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Naive | 1158 | 53774910.490193 | 53772138.304848 | 20964867.878683 | 0.999948 | 0.389863 | 2.564869 | 46477.882878 | 53772138.304848 | 18119.989466 | 134.552504 | 4.563112 | 2004-04-28 | 2004-05-01 | Naive | 73 | 53774910.490193 | 950965.018231 | 4596426.807399 | 0.017684 | 0.085475 | 0.206892 | 46477.882878 | 950965.018231 | 34548.793047 | 250.92778 | 1.586831 | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | 1158 | 53774910.490193 | 53317104.312277 | 20969437.771569 | 0.991487 | 0.389948 | 2.54261 | 46477.882878 | 53317104.312277 | 18123.929093 | 134.567168 | 4.63731 | 2004-04-28 | 2004-05-01 | 73 | 2487513.099357 | 2175054.00871 | 4662501.182581 | 0.874389 | 1.874362 | 0.466499 | 34548.793047 | 2175054.00871 | 34547.877459 | 252.724908 | 1.562477 |
| PT08.S5(O3) | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Naive | 1158 | 111778618.757775 | 111779298.311131 | 25473412.186856 | 1.000006 | 0.227892 | 4.388077 | 96610.733585 | 111779298.311131 | 22016.778022 | 148.316436 | 2.161779 | 2004-04-28 | 2004-05-01 | Naive | 73 | 111778618.757775 | 463604.041014 | 8318074.124279 | 0.004148 | 0.074416 | 0.055735 | 96610.733585 | 463604.041014 | 49276.386463 | 337.559211 | 1.246199 | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | 1158 | 111778618.757775 | 111784511.268927 | 25499165.61918 | 1.000053 | 0.228122 | 4.38385 | 96610.733585 | 111784511.268927 | 22039.016001 | 148.391391 | 2.163942 | 2004-04-28 | 2004-05-01 | 73 | 3547899.825336 | 4769478.210775 | 8317328.740873 | 1.34431 | 2.344296 | 0.573439 | 49276.386463 | 4769478.210775 | 49275.701833 | 337.544086 | 1.24628 |
| T | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Naive | 1158 | 14478.410099 | 14483.396179 | 1150.001763 | 1.000344 | 0.079429 | 12.594238 | 12.513751 | 14483.396179 | 0.993894 | 0.996541 | 3.046411 | 2004-04-28 | 2004-05-01 | Naive | 73 | 14478.410099 | 1228.904351 | 389.831062 | 0.084878 | 0.026925 | 3.152402 | 12.513751 | 1228.904351 | 5.296909 | 2.310877 | 0.847462 | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | 1158 | 14478.410099 | 15329.10426 | 1110.44043 | 1.058756 | 0.076696 | 13.804526 | 12.513751 | 15329.10426 | 0.959753 | 0.979249 | 3.099109 | 2004-04-28 | 2004-05-01 | 73 | 381.377482 | 32.870467 | 410.825195 | 0.086189 | 1.077214 | 0.080011 | 5.296909 | 32.870467 | 5.273289 | 2.372286 | 0.641351 |
| RH | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Naive | 1158 | 160435.204951 | 160336.131853 | 19916.41329 | 0.999382 | 0.12414 | 8.050452 | 138.664827 | 160336.131853 | 17.213529 | 4.147165 | 1.473727 | 2004-04-28 | 2004-05-01 | Naive | 73 | 160435.204951 | 11669.011393 | 29221.10802 | 0.072733 | 0.182137 | 0.399335 | 138.664827 | 11669.011393 | 145.912831 | 20.007227 | 6.707568 | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | 1158 | 160435.204951 | 167371.766979 | 19821.190039 | 1.043236 | 0.123546 | 8.444083 | 138.664827 | 167371.766979 | 17.131429 | 4.137239 | 1.424345 | 2004-04-28 | 2004-05-01 | 73 | 10505.723806 | 17697.983183 | 28158.598116 | 1.684604 | 2.68031 | 0.628511 | 145.912831 | 17697.983183 | 145.29104 | 19.640118 | 6.476134 |
| AH | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Naive | 1158 | 27.198789 | 27.204078 | 1.737284 | 1.000194 | 0.063874 | 15.658966 | 0.023508 | 27.204078 | 0.001502 | 0.038733 | 1.280164 | 2004-04-28 | 2004-05-01 | Naive | 73 | 27.198789 | 0.000389 | 8.432672 | 0.000014 | 0.310039 | 0.000046 | 0.023508 | 0.000389 | 0.029993 | 0.339877 | 1.125849 | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | 1158 | 27.198789 | 27.216369 | 1.736979 | 1.000646 | 0.063862 | 15.668791 | 0.023508 | 27.216369 | 0.001501 | 0.03873 | 1.280324 | 2004-04-28 | 2004-05-01 | 73 | 2.159501 | 6.272988 | 8.432482 | 2.904833 | 3.90483 | 0.743908 | 0.029993 | 6.272988 | 0.029993 | 0.339873 | 1.125817 |
Improvement Model
- Univariate: ARDL, SARIMAX, SVM
- Target(5): 'PT08.S1(CO)', 'PT08.S2(NMHC)', 'PT08.S3(NOx)', 'PT08.S4(NO2)', 'PT08.S5(O3)'
- h-step forecast evaluation
- [Train Period]: 2004-03-10 18:00:00 ~ 2004-04-27 23:00:00, 1158
- [Test Period]: 2004-04-28 00:00:00 ~ 2004-05-01 00:00:00, 73

| start | end | scenario | total | sst | ssr | sse | ssr/sst | sse/sst | ssr/sse | mst | msr | mse | rmse | mape | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| note | domain_kind | model | |||||||||||||||
| PT08.S1(CO) | test | ARDL | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 1.363559e+06 | 1.592328e+06 | 5.298400e+05 | 1.167773 | 0.388571 | 3.005300 | 18938.320225 | 8.380674e+04 | 4157.404298 | 85.194381 | 2.525607 |
| LinearSVR | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 1.363559e+06 | 1.277322e+06 | 7.226992e+04 | 0.936756 | 0.053001 | 17.674321 | 18938.320225 | 4.561863e+04 | 1584.218037 | 31.464248 | 0.780669 | ||
| SARIMAX | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 1.363559e+06 | 1.817311e+06 | 3.533621e+05 | 1.332770 | 0.259147 | 5.142915 | 18938.320225 | 1.397931e+05 | 3133.572945 | 69.574250 | 1.588113 | ||
| train | ARDL | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 3.732967e+07 | 3.285384e+07 | 4.450862e+06 | 0.880100 | 0.119231 | 7.381455 | 32264.189139 | 1.729150e+06 | 3914.566782 | 62.050198 | 1.301650 | |
| LinearSVR | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 3.732967e+07 | 3.588555e+07 | 1.461420e+06 | 0.961315 | 0.039149 | 24.555270 | 32264.189139 | 1.281627e+06 | 1292.377119 | 35.524926 | 0.811643 | ||
| SARIMAX | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 3.732967e+07 | 3.815579e+07 | 1.351412e+06 | 1.022130 | 0.036202 | 28.234027 | 32264.189139 | 2.935061e+06 | 1180.271172 | 34.161705 | 0.759937 | ||
| PT08.S2(NMHC) | test | ARDL | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 1.367597e+06 | 1.628822e+06 | 7.083755e+04 | 1.191011 | 0.051797 | 22.993770 | 18994.399669 | 8.572749e+04 | 1305.730560 | 31.150880 | 0.402409 |
| LinearSVR | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 1.367597e+06 | 1.609306e+06 | 4.022362e+04 | 1.176740 | 0.029412 | 40.008983 | 18994.399669 | 5.747522e+04 | 815.637473 | 23.473569 | 0.284144 | ||
| SARIMAX | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 1.367597e+06 | 1.500905e+06 | 5.445007e+04 | 1.097476 | 0.039814 | 27.564787 | 18994.399669 | 1.154542e+05 | 730.860237 | 27.311012 | 0.347112 | ||
| train | ARDL | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 3.947260e+07 | 3.815931e+07 | 1.214012e+06 | 0.966729 | 0.030756 | 31.432401 | 34116.338096 | 2.008385e+06 | 1067.732492 | 32.406531 | 0.537517 | |
| LinearSVR | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 3.947260e+07 | 4.073622e+07 | 7.153512e+05 | 1.032013 | 0.018123 | 56.945763 | 34116.338096 | 1.454865e+06 | 632.537290 | 24.854521 | 0.431385 | ||
| SARIMAX | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 3.947260e+07 | 3.804603e+07 | 6.851516e+05 | 0.963859 | 0.017358 | 55.529360 | 34116.338096 | 2.926618e+06 | 598.384755 | 24.324228 | 0.363650 | ||
| PT08.S3(NOx) | test | ARDL | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 6.140767e+05 | 9.816835e+05 | 8.655285e+05 | 1.598633 | 1.409479 | 1.134201 | 8528.843129 | 5.166755e+04 | 15413.107054 | 108.887808 | 0.570109 |
| LinearSVR | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 6.140767e+05 | 8.963472e+05 | 2.064890e+05 | 1.459666 | 0.336259 | 4.340896 | 8528.843129 | 3.201240e+04 | 4351.676994 | 53.184736 | 0.270153 | ||
| SARIMAX | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 6.140767e+05 | 8.446576e+05 | 4.360814e+05 | 1.375492 | 0.710142 | 1.936927 | 8528.843129 | 6.033269e+04 | 7267.127889 | 77.289830 | 0.342547 | ||
| train | ARDL | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 4.289261e+07 | 3.592588e+07 | 6.731408e+06 | 0.837577 | 0.156936 | 5.337053 | 37072.268300 | 1.890836e+06 | 5920.323439 | 76.308696 | 3.104840 | |
| LinearSVR | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 4.289261e+07 | 4.306446e+07 | 4.273258e+06 | 1.004006 | 0.099627 | 10.077665 | 37072.268300 | 1.538016e+06 | 3779.092038 | 60.747059 | 2.002940 | ||
| SARIMAX | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 4.289261e+07 | 3.842578e+07 | 4.068667e+06 | 0.895860 | 0.094857 | 9.444315 | 37072.268300 | 2.744698e+06 | 3556.378714 | 59.275035 | 2.714925 | ||
| PT08.S4(NO2) | test | ARDL | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 2.487513e+06 | 2.800260e+06 | 1.393040e+05 | 1.125727 | 0.056001 | 20.101799 | 34548.793047 | 1.473821e+05 | 1366.575486 | 43.683789 | 0.370328 |
| LinearSVR | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 2.487513e+06 | 2.335696e+06 | 5.726143e+04 | 0.938969 | 0.023020 | 40.790050 | 34548.793047 | 8.341773e+04 | 1111.148154 | 28.007197 | 0.257784 | ||
| SARIMAX | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 2.487513e+06 | 2.666173e+06 | 7.631002e+04 | 1.071823 | 0.030677 | 34.938696 | 34548.793047 | 2.050902e+05 | 1157.136276 | 32.331762 | 0.448569 | ||
| train | ARDL | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 5.377491e+07 | 5.202426e+07 | 1.626461e+06 | 0.967445 | 0.030246 | 31.986165 | 46477.882878 | 2.738119e+06 | 1430.484935 | 37.509645 | 5.159292 | |
| LinearSVR | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 5.377491e+07 | 4.907647e+07 | 1.322227e+06 | 0.912628 | 0.024588 | 37.116538 | 46477.882878 | 1.752731e+06 | 1167.876051 | 33.790815 | 2.236627 | ||
| SARIMAX | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 5.377491e+07 | 5.367240e+07 | 1.037607e+06 | 0.998094 | 0.019295 | 51.727075 | 46477.882878 | 4.128646e+06 | 906.165326 | 29.933828 | 2.179873 | ||
| PT08.S5(O3) | test | ARDL | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 3.547900e+06 | 2.988311e+06 | 7.365706e+05 | 0.842276 | 0.207607 | 4.057060 | 49276.386463 | 1.572795e+05 | 13470.632327 | 100.449033 | 0.822711 |
| LinearSVR | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 3.547900e+06 | 3.445754e+06 | 4.627351e+05 | 0.971209 | 0.130425 | 7.446494 | 49276.386463 | 1.230626e+05 | 10274.026451 | 79.616810 | 0.858060 | ||
| SARIMAX | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 3.547900e+06 | 3.258993e+06 | 1.162249e+06 | 0.918570 | 0.327588 | 2.804040 | 49276.386463 | 2.506918e+05 | 14616.319061 | 126.179311 | 1.053004 | ||
| train | ARDL | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 1.117786e+08 | 1.034142e+08 | 8.219973e+06 | 0.925170 | 0.073538 | 12.580850 | 96610.733585 | 5.442855e+06 | 7229.528078 | 84.324988 | 1.084783 | |
| LinearSVR | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 1.117786e+08 | 1.122560e+08 | 4.474445e+06 | 1.004271 | 0.040030 | 25.088249 | 96610.733585 | 4.009142e+06 | 3959.466735 | 62.160615 | 0.726864 | ||
| SARIMAX | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 1.117786e+08 | 1.041239e+08 | 4.617267e+06 | 0.931519 | 0.041307 | 22.550975 | 96610.733585 | 8.009529e+06 | 4031.971021 | 63.144891 | 0.672294 |
- Multivariate: VARMAX[Reduced, Structural-Orthogonal Residual]

| datetime | start | end | scenario | total | sst | ssr | sse | ssr/sst | sse/sst | ssr/sse | mst | msr | mse | rmse | mape | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| note | domain_kind | model | ||||||||||||||||
| PT08.S1(CO) | Train | VAR | 2023-08-28 15:06:01 | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 3.732967e+07 | 3.440836e+07 | 6.167148e+06 | 0.921743 | 0.165208 | 5.579298 | 32264.189139 | 1.810966e+06 | 5414.502498 | 72.977318 | 1.625518 |
| SVAR | 2023-08-28 15:06:01 | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 3.732967e+07 | 3.038897e+07 | 3.844580e+06 | 0.814070 | 0.102990 | 7.904366 | 32264.189139 | 1.599419e+06 | 3374.552238 | 57.619593 | 1.292810 | ||
| Test | VAR | 2023-08-28 15:06:01 | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 1.363559e+06 | 8.841824e+05 | 5.856959e+05 | 0.648437 | 0.429535 | 1.509627 | 18938.320225 | 4.653591e+04 | 9966.437971 | 89.572493 | 2.808808 | |
| SVAR | 2023-08-28 15:06:01 | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 1.363559e+06 | 1.277609e+06 | 3.308650e+05 | 0.936966 | 0.242648 | 3.861420 | 18938.320225 | 6.724256e+04 | 4379.542870 | 67.323083 | 6.351021 | ||
| PT08.S2(NMHC) | Train | VAR | 2023-08-28 15:06:01 | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 3.947260e+07 | 3.688051e+07 | 4.972394e+06 | 0.934332 | 0.125971 | 7.417053 | 34116.338096 | 1.941080e+06 | 4365.500487 | 65.528239 | 1.709778 |
| SVAR | 2023-08-28 15:06:01 | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 3.947260e+07 | 3.553729e+07 | 2.001193e+06 | 0.900303 | 0.050698 | 17.758050 | 34116.338096 | 1.870383e+06 | 1756.407807 | 41.570976 | 0.991264 | ||
| Test | VAR | 2023-08-28 15:06:01 | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 1.367597e+06 | 8.860699e+05 | 5.025864e+05 | 0.647903 | 0.367496 | 1.763020 | 18994.399669 | 4.663526e+04 | 9297.071156 | 82.974368 | 0.803243 | |
| SVAR | 2023-08-28 15:06:01 | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 1.367597e+06 | 1.388727e+06 | 1.974476e+05 | 1.015451 | 0.144376 | 7.033398 | 18994.399669 | 7.309092e+04 | 3360.463086 | 52.007321 | 0.631011 | ||
| PT08.S3(NOx) | Train | VAR | 2023-08-28 15:06:01 | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 4.289261e+07 | 4.000426e+07 | 5.003658e+06 | 0.932661 | 0.116655 | 7.995002 | 37072.268300 | 2.105487e+06 | 4393.006428 | 65.733923 | 1.940422 |
| SVAR | 2023-08-28 15:06:01 | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 4.289261e+07 | 3.838154e+07 | 4.556670e+06 | 0.894829 | 0.106234 | 8.423156 | 37072.268300 | 2.020081e+06 | 4000.048144 | 62.729162 | 1.937093 | ||
| Test | VAR | 2023-08-28 15:06:01 | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 6.140767e+05 | 1.790916e+06 | 1.491800e+06 | 2.916437 | 2.429339 | 1.200507 | 8528.843129 | 9.425875e+04 | 27227.273700 | 142.953215 | 0.726407 | |
| SVAR | 2023-08-28 15:06:01 | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 6.140767e+05 | 2.087912e+06 | 1.445818e+06 | 3.400084 | 2.354459 | 1.444104 | 8528.843129 | 1.098901e+05 | 26371.402302 | 140.732827 | 0.592418 | ||
| PT08.S4(NO2) | Train | VAR | 2023-08-28 15:06:01 | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 5.377491e+07 | 4.580911e+07 | 1.225976e+07 | 0.851868 | 0.227983 | 3.736542 | 46477.882878 | 2.411006e+06 | 10763.456546 | 102.893212 | 2.882381 |
| SVAR | 2023-08-28 15:06:01 | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 5.377491e+07 | 4.531533e+07 | 4.802234e+06 | 0.842685 | 0.089302 | 9.436304 | 46477.882878 | 2.385018e+06 | 4212.798105 | 64.397256 | 10.319381 | ||
| Test | VAR | 2023-08-28 15:06:01 | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 2.487513e+06 | 1.205716e+06 | 1.228865e+06 | 0.484708 | 0.494013 | 0.981163 | 34548.793047 | 6.345875e+04 | 22407.270330 | 129.745001 | 0.739179 | |
| SVAR | 2023-08-28 15:06:01 | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 2.487513e+06 | 2.151442e+06 | 4.136605e+05 | 0.864897 | 0.166295 | 5.200986 | 34548.793047 | 1.132338e+05 | 6242.583281 | 75.276700 | 0.578552 | ||
| PT08.S5(O3) | Train | VAR | 2023-08-28 15:06:01 | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 1.117786e+08 | 1.018719e+08 | 9.551881e+06 | 0.911372 | 0.085454 | 10.665114 | 96610.733585 | 5.361679e+06 | 8386.052792 | 90.821816 | 1.157383 |
| SVAR | 2023-08-28 15:06:01 | 2004-03-10 18:00:00 | 2004-04-27 23:00:00 | Time Series Analysis | 1158 | 1.117786e+08 | 1.158184e+08 | 9.305864e+06 | 1.036141 | 0.083253 | 12.445746 | 96610.733585 | 6.095706e+06 | 8170.008120 | 89.644588 | 1.336284 | ||
| Test | VAR | 2023-08-28 15:06:01 | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 3.547900e+06 | 2.329879e+06 | 1.449389e+06 | 0.656693 | 0.408520 | 1.607490 | 49276.386463 | 1.226252e+05 | 23623.928166 | 140.906524 | 1.434259 | |
| SVAR | 2023-08-28 15:06:01 | 2004-04-28 00:00:00 | 2004-05-01 00:00:00 | Time Series Analysis | 73 | 3.547900e+06 | 7.067373e+06 | 3.579318e+06 | 1.991988 | 1.008855 | 1.974503 | 49276.386463 | 3.719670e+05 | 47682.765355 | 221.431144 | 2.086808 |
Time Series Model Design
Overall Model Specification
Baseline model
Naive forecast model
- endogenous variables: 'PT08.S1', 'PT08.S2', 'PT08.S3', 'PT08.S4', 'PT08.S5', 'CO(GT)', 'NMHC(GT)', 'C6H6(GT)', 'NOx(GT)', 'NO2(GT)', 'T', 'RH', 'AH'
- [Train Period]: 2004-03-10 18:00:00 ~ 2004-04-27 23:00:00, 1158
- [Test Period]: 2004-04-28 00:00:00 ~ 2004-05-01 00:00:00, 73
ETS model: Addictive Dampping Trend
- endogenous variables: 'PT08.S1', 'PT08.S2', 'PT08.S3', 'PT08.S4', 'PT08.S5', 'CO(GT)', 'NMHC(GT)', 'C6H6(GT)', 'NOx(GT)', 'NO2(GT)', 'T', 'RH', 'AH'
- [Train Period]: 2004-03-10 18:00:00 ~ 2004-04-27 23:00:00, 1158
- [Test Period]: 2004-04-28 00:00:00 ~ 2004-05-01 00:00:00, 73
Improvement model
Short-term forecast model
- Univariate approach
- ARDL: Autoregressive Distributed Lag Model
- endogenous variables[-2 lag]: 'PT08.S1', 'PT08.S2', 'PT08.S3', 'PT08.S4', 'PT08.S5'
- exogenous variables[-2 lag]: 'CO(GT)', 'NMHC(GT)', 'C6H6(GT)', 'NOx(GT)', 'NO2(GT)', 'T', 'RH', 'AH'
- bias: constant
- [Train Period]: 2004-03-10 18:00:00 ~ 2004-04-27 23:00:00, 1158
- [Test Period]: 2004-04-28 00:00:00 ~ 2004-05-01 00:00:00, 73
- (note) Stationary Constraint[False], Integration Structure[False], Error Correction[False], Distributed Lag[True]
- SARIMAX: Autoregressive Moving Average Model
- endogenous variables[-1,-2 lag]: 'PT08.S1', 'PT08.S2', 'PT08.S3', 'PT08.S4', 'PT08.S5'
- exogenous variables: 'CO(GT)', 'NMHC(GT)', 'C6H6(GT)', 'NOx(GT)', 'NO2(GT)', 'T', 'RH', 'AH'
- bias: constant
- constant volatility: -1, -2 lag
- [Train Period]: 2004-03-10 18:00:00 ~ 2004-04-27 23:00:00, 1158
- [Test Period]: 2004-04-28 00:00:00 ~ 2004-05-01 00:00:00, 73
- (note) Stationary Constraint[True], Integration Structure[True], Error Correction[False], Distributed Lag[False]
- SVM: Support Vector Machine
- endogenous variables[-2lag]: 'PT08.S1', 'PT08.S2', 'PT08.S3', 'PT08.S4', 'PT08.S5'
- fixed exogenous variables[-2lag]: 'CO(GT)', 'NMHC(GT)', 'C6H6(GT)', 'NOx(GT)', 'NO2(GT)', 'T', 'RH', 'AH'
- bias: constant
- [Train Period]: 2004-03-10 18:00:00 ~ 2004-04-27 23:00:00, 1158
- [Test Period]: 2004-04-28 00:00:00 ~ 2004-05-01 00:00:00, 73
- (note) Stationary Constraint[False], Integration Structure[True], Error Correction[True], Distributed Lag[True]
- ARDL: Autoregressive Distributed Lag Model
- Multivariate approach
- VARMAX: Vector Autoregressive Moving Average Model
- endogenous variables[-2lag]: 'PT08.S1', 'PT08.S2', 'PT08.S3', 'PT08.S4', 'PT08.S5'
- exogenous variables: 'CO(GT)', 'NMHC(GT)', 'C6H6(GT)', 'NOx(GT)', 'NO2(GT)', 'T', 'RH', 'AH'
- bias: constant
- [Train Period]: 2004-03-10 18:00:00 ~ 2004-04-27 23:00:00, 1158
- [Test Period]: 2004-04-28 00:00:00 ~ 2004-05-01 00:00:00, 73
- (note) Stationary Constraint[True], Integration Structure[False], Constraint Error Correction[False], Distributed Lag[False]
- VECM: Vector Error Correction Model
- endogenous variables[-2lag]: 'PT08.S1', 'PT08.S2', 'PT08.S3', 'PT08.S4', 'PT08.S5'
- exogenous variables: 'T', 'RH', 'AH'
- coint exogenous variables[-lag]: 'CO(GT)', 'NMHC(GT)', 'C6H6(GT)', 'NOx(GT)', 'NO2(GT)'
- bias: constant
- cointegration rank: 4
- [Train Period]: 2004-03-10 18:00:00 ~ 2004-04-27 23:00:00, 1158
- [Test Period]: 2004-04-28 00:00:00 ~ 2004-05-01 00:00:00, 73
- (note) Stationary Constraint[True], Integration Structure[True], Constraint Error Correction[True], Distributed Lag[False]
- VARMAX: Vector Autoregressive Moving Average Model
Base Preprocessing for Forecasting Pollutants Variables
- Statistical characteristics preprocessing: mean-centering[False], variance-centering[False]
- Time Series characteristics preprocessing: lag shift[True], trend difference[True], seasonal difference[False]
Multivariate Time Series Characteristic Analysis
[Note] Dynamic Facot Analysis (DFA), Principal Component Analysis (PCA), and Cointegration Analysis were conducted on deseasonalized time series data.
Dynamic Factor Analysis
Dynamic Factor Analysis (DFA) uncovers common factors that can explain multiple given time series data collectively. This method aims to go beyond individual approachs for each time series data and seeks integrated insights by understanding the interrelationships among them. Also, DFA involves reducing the dimensions of the given multidimensional data while explaining the variablility in the data by extracting significant dynamic factors. These factors can reveal shared structures or relationships among the given time series data. Consequently, it helps to identify patterns, similarities, and correlations among the data, providing a comprehensive overview.
Dynamic factor analysis can be seen as an extended version of Principal Component Analysis (PCA) applied to time series data. While PCA primarily focuses on extracting principal components that explain the variability in the data, dynamic factor analysis emphasizes extracting factors while considering the temporal structure of the time series data. This approach allows us to understand interactions and dynamic relationships among multiple time series data, shedding light on how these data sets mutually influence each other.
| coef | std err | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| L1.f1.f1 | 0.0489 | 0.027 | 1.779 | 0.075 | -0.005 | 0.103 |
| L2.f1.f1 | -0.2070 | 0.031 | -6.708 | 0.000 | -0.268 | -0.147 |




Result for factor analysis
| First Factor Common Loadings | Unique Loadings Variance | Lag1 | Lag2 | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| coef | std err | z | P>|z| | [0.025 | 0.975] | coef | std err | z | P>|z| | [0.025 | 0.975] | coef | std err | z | P>|z| | [0.025 | 0.975] | coef | std err | z | P>|z| | [0.025 | 0.975] | |||||
| CO(GT) | loading.f1.CO(GT) | -0.5623 | 0.018 | -30.888 | 0.000 | -0.598 | -0.527 | sigma2.CO(GT) | 0.1611 | 0.007 | 23.919 | 0.000 | 0.148 | 0.174 | L1.e(CO(GT)).e(CO(GT)) | -0.1209 | 0.028 | -4.330 | 0.000 | -0.176 | -0.066 | L2.e(CO(GT)).e(CO(GT)) | -0.2414 | 0.032 | -7.539 | 0.000 | -0.304 | -0.179 |
| PT08.S1(CO) | loading.f1.PT08.S1(CO) | -0.5346 | 0.012 | -44.319 | 0.000 | -0.558 | -0.511 | sigma2.PT08.S1(CO) | 0.0341 | 0.001 | 23.153 | 0.000 | 0.031 | 0.037 | L1.e(PT08.S1(CO)).e(PT08.S1(CO)) | -0.2784 | 0.029 | -9.497 | 0.000 | -0.336 | -0.221 | L2.e(PT08.S1(CO)).e(PT08.S1(CO)) | -0.1702 | 0.033 | -5.159 | 0.000 | -0.235 | -0.106 |
| NMHC(GT) | loading.f1.NMHC(GT) | -0.4683 | 0.018 | -25.642 | 0.000 | -0.504 | -0.432 | sigma2.NMHC(GT) | 0.2610 | 0.008 | 31.868 | 0.000 | 0.245 | 0.277 | L1.e(NMHC(GT)).e(NMHC(GT)) | -0.0974 | 0.025 | -3.916 | 0.000 | -0.146 | -0.049 | L2.e(NMHC(GT)).e(NMHC(GT)) | -0.2291 | 0.029 | -7.808 | 0.000 | -0.287 | -0.172 |
| C6H6(GT) | loading.f1.C6H6(GT) | -0.6151 | 0.015 | -41.809 | 0.000 | -0.644 | -0.586 | sigma2.C6H6(GT) | 0.0219 | 0.001 | 19.103 | 0.000 | 0.020 | 0.024 | L1.e(C6H6(GT)).e(C6H6(GT)) | -0.1276 | 0.034 | -3.755 | 0.000 | -0.194 | -0.061 | L2.e(C6H6(GT)).e(C6H6(GT)) | -0.1920 | 0.045 | -4.289 | 0.000 | -0.280 | -0.104 |
| PT08.S2(NMHC) | loading.f1.PT08.S2(NMHC) | -0.5711 | 0.012 | -48.124 | 0.000 | -0.594 | -0.548 | sigma2.PT08.S2(NMHC) | 0.0072 | 0.001 | 9.625 | 0.000 | 0.006 | 0.009 | L1.e(PT08.S2(NMHC)).e(PT08.S2(NMHC)) | 0.3806 | 0.067 | 5.675 | 0.000 | 0.249 | 0.512 | L2.e(PT08.S2(NMHC)).e(PT08.S2(NMHC)) | -0.3231 | 0.055 | -5.823 | 0.000 | -0.432 | -0.214 |
| NOx(GT) | loading.f1.NOx(GT) | -0.4926 | 0.022 | -22.761 | 0.000 | -0.535 | -0.450 | sigma2.NOx(GT) | 0.1989 | 0.009 | 22.596 | 0.000 | 0.182 | 0.216 | L1.e(NOx(GT)).e(NOx(GT)) | -0.1743 | 0.030 | -5.816 | 0.000 | -0.233 | -0.116 | L2.e(NOx(GT)).e(NOx(GT)) | -0.2059 | 0.032 | -6.467 | 0.000 | -0.268 | -0.143 |
| PT08.S3(NOx) | loading.f1.PT08.S3(NOx) | 0.3863 | 0.020 | 19.610 | 0.000 | 0.348 | 0.425 | sigma2.PT08.S3(NOx) | 0.0846 | 0.003 | 29.112 | 0.000 | 0.079 | 0.090 | L1.e(PT08.S3(NOx)).e(PT08.S3(NOx)) | 0.1887 | 0.024 | 7.765 | 0.000 | 0.141 | 0.236 | L2.e(PT08.S3(NOx)).e(PT08.S3(NOx)) | -0.2037 | 0.027 | -7.586 | 0.000 | -0.256 | -0.151 |
| NO2(GT) | loading.f1.NO2(GT) | -0.3610 | 0.021 | -16.892 | 0.000 | -0.403 | -0.319 | sigma2.NO2(GT) | 0.2572 | 0.011 | 23.704 | 0.000 | 0.236 | 0.278 | L1.e(NO2(GT)).e(NO2(GT)) | -0.1683 | 0.025 | -6.684 | 0.000 | -0.218 | -0.119 | L2.e(NO2(GT)).e(NO2(GT)) | -0.2548 | 0.028 | -9.033 | 0.000 | -0.310 | -0.200 |
| PT08.S4(NO2) | loading.f1.PT08.S4(NO2) | -0.5643 | 0.012 | -45.571 | 0.000 | -0.589 | -0.540 | sigma2.PT08.S4(NO2) | 0.0219 | 0.000 | 59.972 | 0.000 | 0.021 | 0.023 | L1.e(PT08.S4(NO2)).e(PT08.S4(NO2)) | -0.5146 | 0.013 | -39.464 | 0.000 | -0.540 | -0.489 | L2.e(PT08.S4(NO2)).e(PT08.S4(NO2)) | -0.2518 | 0.024 | -10.355 | 0.000 | -0.299 | -0.204 |
| PT08.S5(O3) | loading.f1.PT08.S5(O3) | -0.4193 | 0.010 | -40.343 | 0.000 | -0.440 | -0.399 | sigma2.PT08.S5(O3) | 0.0413 | 0.001 | 28.347 | 0.000 | 0.038 | 0.044 | L1.e(PT08.S5(O3)).e(PT08.S5(O3)) | 0.0484 | 0.027 | 1.781 | 0.075 | -0.005 | 0.102 | L2.e(PT08.S5(O3)).e(PT08.S5(O3)) | -0.1730 | 0.032 | -5.397 | 0.000 | -0.236 | -0.110 |
| T | loading.f1.T | 0.0041 | 0.019 | 0.212 | 0.832 | -0.033 | 0.042 | sigma2.T | 0.0731 | 0.004 | 20.496 | 0.000 | 0.066 | 0.080 | L1.e(T).e(T) | 0.2739 | 0.034 | 8.042 | 0.000 | 0.207 | 0.341 | L2.e(T).e(T) | -0.0180 | 0.038 | -0.472 | 0.637 | -0.093 | 0.057 |
| RH | loading.f1.RH | -0.0403 | 0.032 | -1.257 | 0.209 | -0.103 | 0.022 | sigma2.RH | 0.1186 | 0.005 | 24.486 | 0.000 | 0.109 | 0.128 | L1.e(RH).e(RH) | 0.2269 | 0.029 | 7.957 | 0.000 | 0.171 | 0.283 | L2.e(RH).e(RH) | -0.0631 | 0.034 | -1.876 | 0.061 | -0.129 | 0.003 |
| AH | loading.f1.AH | -0.0488 | 0.014 | -3.496 | 0.000 | -0.076 | -0.021 | sigma2.AH | 0.0505 | 0.001 | 33.690 | 0.000 | 0.048 | 0.053 | L1.e(AH).e(AH) | 0.1954 | 0.021 | 9.206 | 0.000 | 0.154 | 0.237 | L2.e(AH).e(AH) | -0.1380 | 0.027 | -5.180 | 0.000 | -0.190 | -0.086 |
Diagnostics for residual
- There exists a limitation of non-conformity to normality for the residuals. This stems from training with only one extracted factor. It is anticipated that an enhancement could be achieved by increasing the number of factors.

| Autocorrelation | Heteroskedasticity | Normality | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ljung-Box (L1) (Q) | Prob(Q) | Reject | Heteroskedasticity (H) | Prob(H) (two-sided) | Reject | Jarque-Bera (JB) | Prob(JB) | Skew | Kurtosis | Reject | |
| Residual | |||||||||||
| CO(GT) | 0.62 | 0.43 | False | 0.64 | 0.00 | True | 762.43 | 0.0 | 0.41 | 6.77 | False |
| PT08.S1(CO) | 0.00 | 0.99 | False | 1.96 | 0.00 | True | 385.30 | 0.0 | 0.25 | 5.70 | False |
| NMHC(GT) | 2.44 | 0.12 | False | 1.27 | 0.02 | True | 979.78 | 0.0 | 0.23 | 7.35 | False |
| C6H6(GT) | 0.45 | 0.50 | False | 1.29 | 0.01 | True | 1584.67 | 0.0 | 0.05 | 8.56 | False |
| PT08.S2(NMHC) | 5.72 | 0.02 | True | 0.91 | 0.34 | False | 36.53 | 0.0 | -0.08 | 3.83 | False |
| NOx(GT) | 0.81 | 0.37 | False | 1.19 | 0.08 | False | 519.95 | 0.0 | -0.07 | 6.18 | False |
| PT08.S3(NOx) | 2.13 | 0.14 | False | 0.74 | 0.00 | True | 502.91 | 0.0 | 0.10 | 6.13 | False |
| NO2(GT) | 2.59 | 0.11 | False | 0.77 | 0.01 | True | 59.82 | 0.0 | 0.08 | 4.07 | False |
| PT08.S4(NO2) | 0.05 | 0.83 | False | 1.12 | 0.27 | False | 198431.95 | 0.0 | -2.34 | 65.05 | False |
| PT08.S5(O3) | 0.46 | 0.50 | False | 0.85 | 0.10 | False | 356.53 | 0.0 | 0.46 | 5.48 | False |
| T | 0.00 | 0.99 | False | 1.21 | 0.05 | False | 708.67 | 0.0 | -0.27 | 6.68 | False |
| RH | 0.00 | 1.00 | False | 1.03 | 0.74 | False | 3498.46 | 0.0 | 0.56 | 11.19 | False |
| AH | 0.03 | 0.87 | False | 1.31 | 0.01 | True | 2153.34 | 0.0 | 0.71 | 9.32 | False |
Principal Component Analysis
Principal Component Analysis (PCA) is a widely used statistical technique for analyzing and reducing the dimensions of multivariate data. It aims to capture the most important information or patterns in the data while discarding or minimizing the less important information. PCA achieves this by transforming the original variables into a new set of uncorrelated variables called principal components.


Cointegration Analysis

Bivariate cointegration Analysis
cointegration for deseasonalized time series
- [PT08.S1(CO)]: PT08.S2(NMHC), NOx(GT), NO2(GT), CO(GT)
- [PT08.S2(NMHC)]: PT08.S1(CO), PT08.S4(NO2), PT08.S5(O3), CO(GT), C6H6(GT), T, AH
- [PT08.S3(NOx)]: PT08.S4(NO2), PT08.S5(O3), CO(GT), NMHC(GT), C6H6(GT), NOx(GT), NO2(GT), T, RH, AH
- [PT08.S4(NO2)]: PT08.S2(NMHC), PT08.S3(NOx), C6H6(GT)
- [PT08.S5(O3)]: PT08.S2(NMHC), PT08.S3(NOx), CO(GT), C6H6(GT), NOx(GT), NO2(GT), AH
| Cointegration | RV1ResidualStationary | RV2ResidualStationary | Cointegration | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rv1 | rv2 | t-statistic | p-val | crit1 | crit5 | crit10 | t-statistic | p-val | used_lag | nobs | crit1 | crit5 | crit10 | icbest | t-statistic | p-val | used_lag | nobs | crit1 | crit5 | crit10 | icbest | Determination | |
| 0 | CO(GT) | PT08.S1(CO) | -7.295834 | 1.667493e-09 | -3.905366 | -3.341102 | -3.0479 | -7.294736 | 1.387455e-10 | 8 | 1222 | -3.435713 | -2.863908 | -2.568031 | 1046.851611 | -3.519355 | 7.503711e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 13444.258432 | True |
| 1 | CO(GT) | PT08.S2(NMHC) | -3.747276 | 1.591290e-02 | -3.905366 | -3.341102 | -3.0479 | -3.745636 | 3.515157e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 1030.167743 | -3.264312 | 1.655105e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 13596.564121 | True |
| 2 | CO(GT) | NOx(GT) | -4.429011 | 1.598218e-03 | -3.905366 | -3.341102 | -3.0479 | -4.425954 | 2.669638e-04 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 328.344393 | -4.433306 | 2.590360e-04 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 10181.944235 | True |
| 3 | CO(GT) | PT08.S3(NOx) | -3.987745 | 7.475540e-03 | -3.905366 | -3.341102 | -3.0479 | -3.990248 | 1.462444e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 1695.232821 | -3.926294 | 1.849637e-03 | 22 | 1208 | -3.435775 | -2.863936 | -2.568045 | 14060.499323 | True |
| 4 | CO(GT) | NO2(GT) | -4.342215 | 2.198459e-03 | -3.905366 | -3.341102 | -3.0479 | -4.332872 | 3.894570e-04 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 1484.290900 | -4.046021 | 1.187804e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 8915.923324 | True |
| 5 | CO(GT) | PT08.S5(O3) | -4.837217 | 3.242708e-04 | -3.905366 | -3.341102 | -3.0479 | -4.836122 | 4.636230e-05 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 1309.219236 | -3.808615 | 2.820391e-03 | 22 | 1208 | -3.435775 | -2.863936 | -2.568045 | 14895.258012 | True |
| 6 | CO(GT) | T | -3.451260 | 3.696768e-02 | -3.905366 | -3.341102 | -3.0479 | -3.448194 | 9.421624e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 2177.922142 | -3.369108 | 1.205815e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 3541.283688 | True |
| 7 | CO(GT) | RH | -3.410705 | 4.117532e-02 | -3.905366 | -3.341102 | -3.0479 | -3.408693 | 1.066606e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 2173.113060 | -7.000788 | 7.331272e-10 | 7 | 1223 | -3.435708 | -2.863906 | -2.568030 | 6784.747928 | True |
| 8 | CO(GT) | AH | -3.950840 | 8.428314e-03 | -3.905366 | -3.341102 | -3.0479 | -3.950521 | 1.692950e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 2157.102440 | -3.595769 | 5.842435e-03 | 9 | 1221 | -3.435717 | -2.863910 | -2.568032 | -3847.679236 | True |
| 9 | PT08.S1(CO) | PT08.S2(NMHC) | -3.662927 | 2.043521e-02 | -3.905366 | -3.341102 | -3.0479 | -3.669221 | 4.567518e-03 | 18 | 1212 | -3.435757 | -2.863928 | -2.568041 | 12173.907550 | -5.201615 | 8.725575e-06 | 8 | 1222 | -3.435713 | -2.863908 | -2.568031 | 12354.828445 | True |
| 10 | PT08.S1(CO) | NOx(GT) | -3.399306 | 4.242766e-02 | -3.905366 | -3.341102 | -3.0479 | -3.398576 | 1.100751e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 13616.376740 | -6.889800 | 1.364878e-09 | 9 | 1221 | -3.435717 | -2.863910 | -2.568032 | 11026.449562 | True |
| 11 | PT08.S1(CO) | NO2(GT) | -3.454223 | 3.667491e-02 | -3.905366 | -3.341102 | -3.0479 | -3.448264 | 9.419542e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 14049.379278 | -7.348524 | 1.020419e-10 | 9 | 1221 | -3.435717 | -2.863910 | -2.568032 | 8991.692280 | True |
| 12 | NMHC(GT) | PT08.S3(NOx) | -14.390939 | 7.974083e-26 | -3.905366 | -3.341102 | -3.0479 | -14.385185 | 9.014536e-27 | 2 | 1228 | -3.435686 | -2.863896 | -2.568025 | 14059.837908 | -8.318992 | 3.615332e-13 | 5 | 1225 | -3.435699 | -2.863902 | -2.568028 | 14304.029751 | True |
| 13 | NMHC(GT) | T | -3.925943 | 9.131333e-03 | -3.905366 | -3.341102 | -3.0479 | -3.926583 | 1.847689e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 14329.985341 | -3.137825 | 2.387766e-02 | 22 | 1208 | -3.435775 | -2.863936 | -2.568045 | 3803.168754 | True |
| 14 | NMHC(GT) | RH | -3.733993 | 1.656077e-02 | -3.905366 | -3.341102 | -3.0479 | -3.737017 | 3.621601e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 14335.610189 | -7.995953 | 2.404064e-12 | 1 | 1229 | -3.435682 | -2.863895 | -2.568024 | 6769.329755 | True |
| 15 | NMHC(GT) | AH | -4.627549 | 7.500186e-04 | -3.905366 | -3.341102 | -3.0479 | -4.631431 | 1.129930e-04 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 14327.370198 | -4.937199 | 2.950334e-05 | 2 | 1228 | -3.435686 | -2.863896 | -2.568025 | -3754.712726 | True |
| 16 | C6H6(GT) | PT08.S2(NMHC) | -4.494821 | 1.248891e-03 | -3.905366 | -3.341102 | -3.0479 | -4.492322 | 2.030179e-04 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 2991.887523 | -4.242326 | 5.582399e-04 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 11458.551574 | True |
| 17 | C6H6(GT) | PT08.S3(NOx) | -3.341117 | 4.932446e-02 | -3.905366 | -3.341102 | -3.0479 | -3.345402 | 1.296702e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 5465.016999 | -3.746829 | 3.500654e-03 | 22 | 1208 | -3.435775 | -2.863936 | -2.568045 | 13711.805397 | True |
| 18 | C6H6(GT) | PT08.S4(NO2) | -3.390534 | 4.341279e-02 | -3.905366 | -3.341102 | -3.0479 | -3.385736 | 1.145485e-02 | 19 | 1211 | -3.435761 | -2.863930 | -2.568042 | 3490.381272 | -6.436550 | 1.647404e-08 | 2 | 1228 | -3.435686 | -2.863896 | -2.568025 | 12228.558737 | True |
| 19 | C6H6(GT) | PT08.S5(O3) | -3.980070 | 7.665274e-03 | -3.905366 | -3.341102 | -3.0479 | -3.980435 | 1.516499e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 4814.847207 | -3.429492 | 9.993647e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 14281.517761 | True |
| 20 | C6H6(GT) | T | -3.420487 | 4.012542e-02 | -3.905366 | -3.341102 | -3.0479 | -3.417969 | 1.036134e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 6231.506142 | -3.466463 | 8.891342e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 3884.010504 | True |
| 21 | C6H6(GT) | AH | -3.403249 | 4.199088e-02 | -3.905366 | -3.341102 | -3.0479 | -3.403418 | 1.084289e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 6218.091602 | -3.867152 | 2.290341e-03 | 7 | 1223 | -3.435708 | -2.863906 | -2.568030 | -4095.951931 | True |
| 22 | PT08.S2(NMHC) | PT08.S4(NO2) | -3.646655 | 2.142610e-02 | -3.905366 | -3.341102 | -3.0479 | -3.643185 | 4.987105e-03 | 19 | 1211 | -3.435761 | -2.863930 | -2.568042 | 11955.661649 | -4.772569 | 6.135271e-05 | 9 | 1221 | -3.435717 | -2.863910 | -2.568032 | 12424.244492 | True |
| 23 | PT08.S2(NMHC) | PT08.S5(O3) | -4.001632 | 7.142924e-03 | -3.905366 | -3.341102 | -3.0479 | -4.002383 | 1.398082e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 12796.766940 | -3.628374 | 5.241185e-03 | 22 | 1208 | -3.435775 | -2.863936 | -2.568045 | 13798.462833 | True |
| 24 | PT08.S2(NMHC) | T | -3.409912 | 4.126141e-02 | -3.905366 | -3.341102 | -3.0479 | -3.407089 | 1.071955e-02 | 22 | 1208 | -3.435775 | -2.863936 | -2.568045 | 14601.280067 | -3.479724 | 8.523358e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 3812.854867 | True |
| 25 | PT08.S2(NMHC) | AH | -3.497560 | 3.261231e-02 | -3.905366 | -3.341102 | -3.0479 | -3.497576 | 8.049566e-03 | 22 | 1208 | -3.435775 | -2.863936 | -2.568045 | 14561.669841 | -3.993126 | 1.446935e-03 | 7 | 1223 | -3.435708 | -2.863906 | -2.568030 | -4102.051060 | True |
| 26 | NOx(GT) | PT08.S3(NOx) | -3.972449 | 7.857974e-03 | -3.905366 | -3.341102 | -3.0479 | -3.976581 | 1.538230e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 11507.299261 | -3.929957 | 1.825111e-03 | 21 | 1209 | -3.435770 | -2.863934 | -2.568044 | 13994.311970 | True |
| 27 | NOx(GT) | NO2(GT) | -14.440195 | 6.635408e-26 | -3.905366 | -3.341102 | -3.0479 | -14.434119 | 7.545213e-27 | 2 | 1228 | -3.435686 | -2.863896 | -2.568025 | 10839.311326 | -12.762040 | 8.104430e-24 | 2 | 1228 | -3.435686 | -2.863896 | -2.568025 | 8397.529904 | True |
| 28 | NOx(GT) | PT08.S5(O3) | -4.818442 | 3.501191e-04 | -3.905366 | -3.341102 | -3.0479 | -4.814731 | 5.096475e-05 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 11137.117050 | -3.759384 | 3.351300e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 14863.349608 | True |
| 29 | NOx(GT) | T | -3.468602 | 3.528209e-02 | -3.905366 | -3.341102 | -3.0479 | -3.466439 | 8.892032e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 12001.623297 | -3.370376 | 1.201117e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 3420.532654 | True |
| 30 | NOx(GT) | RH | -3.484945 | 3.375349e-02 | -3.905366 | -3.341102 | -3.0479 | -3.483434 | 8.422877e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 11982.667303 | -8.016872 | 2.127254e-12 | 1 | 1229 | -3.435682 | -2.863895 | -2.568024 | 6772.603716 | True |
| 31 | NOx(GT) | AH | -3.789475 | 1.400009e-02 | -3.905366 | -3.341102 | -3.0479 | -3.788043 | 3.032045e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 11975.148088 | -4.406412 | 2.891716e-04 | 4 | 1226 | -3.435695 | -2.863900 | -2.568027 | -4031.305638 | True |
| 32 | PT08.S3(NOx) | NO2(GT) | -3.454900 | 3.660838e-02 | -3.905366 | -3.341102 | -3.0479 | -3.465596 | 8.915905e-03 | 22 | 1208 | -3.435775 | -2.863936 | -2.568045 | 13975.871802 | -3.564677 | 6.473374e-03 | 22 | 1208 | -3.435775 | -2.863936 | -2.568045 | 9046.197972 | True |
| 33 | PT08.S3(NOx) | PT08.S4(NO2) | -4.226399 | 3.325655e-03 | -3.905366 | -3.341102 | -3.0479 | -4.252339 | 5.366432e-04 | 22 | 1208 | -3.435775 | -2.863936 | -2.568045 | 13506.666744 | -3.021526 | 3.293246e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 14079.554752 | True |
| 34 | PT08.S3(NOx) | PT08.S5(O3) | -3.491750 | 3.313379e-02 | -3.905366 | -3.341102 | -3.0479 | -3.517830 | 7.540815e-03 | 22 | 1208 | -3.435775 | -2.863936 | -2.568045 | 13050.543689 | -2.938197 | 4.108311e-02 | 21 | 1209 | -3.435770 | -2.863934 | -2.568044 | 14162.528573 | True |
| 35 | PT08.S3(NOx) | T | -3.781301 | 1.435389e-02 | -3.905366 | -3.341102 | -3.0479 | -3.783663 | 3.078961e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 14370.577284 | -3.681439 | 4.381935e-03 | 17 | 1213 | -3.435752 | -2.863926 | -2.568040 | 3714.912190 | True |
| 36 | PT08.S3(NOx) | RH | -3.620887 | 2.308052e-02 | -3.905366 | -3.341102 | -3.0479 | -3.628257 | 5.243237e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 14253.920601 | -6.774255 | 2.594867e-09 | 7 | 1223 | -3.435708 | -2.863906 | -2.568030 | 6701.073556 | True |
| 37 | PT08.S3(NOx) | AH | -4.613064 | 7.935735e-04 | -3.905366 | -3.341102 | -3.0479 | -4.622658 | 1.173022e-04 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 14193.389584 | -4.757715 | 6.547430e-05 | 6 | 1224 | -3.435704 | -2.863904 | -2.568029 | -3926.969981 | True |
| 38 | NO2(GT) | PT08.S5(O3) | -11.498055 | 5.446771e-20 | -3.905366 | -3.341102 | -3.0479 | -11.493240 | 4.680783e-21 | 2 | 1228 | -3.435686 | -2.863896 | -2.568025 | 8850.883921 | -7.237153 | 1.926207e-10 | 5 | 1225 | -3.435699 | -2.863902 | -2.568028 | 15060.203145 | True |
| 39 | NO2(GT) | T | -8.285076 | 5.769871e-12 | -3.905366 | -3.341102 | -3.0479 | -8.281688 | 4.501830e-13 | 4 | 1226 | -3.435695 | -2.863900 | -2.568027 | 9520.612251 | -3.369836 | 1.203118e-02 | 20 | 1210 | -3.435766 | -2.863932 | -2.568043 | 3489.442167 | True |
| 40 | NO2(GT) | RH | -8.118876 | 1.517547e-11 | -3.905366 | -3.341102 | -3.0479 | -8.115571 | 1.193561e-12 | 4 | 1226 | -3.435695 | -2.863900 | -2.568027 | 9518.794113 | -8.020461 | 2.083066e-12 | 1 | 1229 | -3.435682 | -2.863895 | -2.568024 | 6762.194011 | True |
| 41 | NO2(GT) | AH | -8.265363 | 6.472569e-12 | -3.905366 | -3.341102 | -3.0479 | -8.261984 | 5.054482e-13 | 4 | 1226 | -3.435695 | -2.863900 | -2.568027 | 9504.021513 | -4.276464 | 4.877980e-04 | 3 | 1227 | -3.435691 | -2.863898 | -2.568026 | -4257.468169 | True |
| 42 | PT08.S5(O3) | AH | -6.226827 | 5.179663e-07 | -3.905366 | -3.341102 | -3.0479 | -6.223971 | 5.142080e-08 | 6 | 1224 | -3.435704 | -2.863904 | -2.568029 | 15311.512143 | -3.776070 | 3.161858e-03 | 8 | 1222 | -3.435713 | -2.863908 | -2.568031 | -4323.111834 | True |
| 43 | T | AH | -3.875211 | 1.072864e-02 | -3.905366 | -3.341102 | -3.0479 | -3.875754 | 2.220712e-03 | 17 | 1213 | -3.435752 | -2.863926 | -2.568040 | 3490.488239 | -5.232609 | 7.539782e-06 | 2 | 1228 | -3.435686 | -2.863896 | -2.568025 | -4122.831833 | True |
| 44 | RH | AH | -7.467609 | 6.350136e-10 | -3.905366 | -3.341102 | -3.0479 | -7.464555 | 5.246078e-11 | 1 | 1229 | -3.435682 | -2.863895 | -2.568024 | 6272.058101 | -3.530808 | 7.230239e-03 | 2 | 1228 | -3.435686 | -2.863896 | -2.568025 | -5020.681117 | True |
| 45 | CO(GT) | NMHC(GT) | -1.855580 | 6.023538e-01 | -3.905366 | -3.341102 | -3.0479 | -1.852638 | 3.546590e-01 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 873.688074 | -2.392712 | 1.438040e-01 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 13055.842895 | False |
| 46 | CO(GT) | C6H6(GT) | -3.055619 | 9.756910e-02 | -3.905366 | -3.341102 | -3.0479 | -3.054041 | 3.014665e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 898.938741 | -2.726789 | 6.951626e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 5018.752458 | False |
| 47 | CO(GT) | PT08.S4(NO2) | -3.798390 | 1.362297e-02 | -3.905366 | -3.341102 | -3.0479 | -3.794999 | 2.958894e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 1087.193383 | -2.571166 | 9.910429e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 14115.501888 | False |
| 48 | PT08.S1(CO) | NMHC(GT) | -2.101053 | 4.758168e-01 | -3.905366 | -3.341102 | -3.0479 | -2.103031 | 2.432977e-01 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 13971.368967 | -3.254258 | 1.705114e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 13847.832224 | False |
| 49 | PT08.S1(CO) | C6H6(GT) | -3.237862 | 6.382093e-02 | -3.905366 | -3.341102 | -3.0479 | -3.245376 | 1.750385e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 12533.665247 | -3.511271 | 7.702305e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 4399.520094 | False |
| 50 | PT08.S1(CO) | PT08.S3(NOx) | -2.991350 | 1.122760e-01 | -3.905366 | -3.341102 | -3.0479 | -3.017573 | 3.328555e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 13445.216485 | -3.919083 | 1.898807e-03 | 21 | 1209 | -3.435770 | -2.863934 | -2.568044 | 13267.115506 | False |
| 51 | PT08.S1(CO) | PT08.S4(NO2) | -2.585475 | 2.427257e-01 | -3.905366 | -3.341102 | -3.0479 | -2.591492 | 9.476848e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 12295.088400 | -2.530594 | 1.082067e-01 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 12949.708352 | False |
| 52 | PT08.S1(CO) | PT08.S5(O3) | -3.325953 | 5.126612e-02 | -3.905366 | -3.341102 | -3.0479 | -3.334911 | 1.338829e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 12672.410974 | -3.747587 | 3.491474e-03 | 22 | 1208 | -3.435775 | -2.863936 | -2.568045 | 13724.672889 | False |
| 53 | PT08.S1(CO) | T | -2.682265 | 2.060221e-01 | -3.905366 | -3.341102 | -3.0479 | -2.681176 | 7.735272e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 14471.240146 | -3.442370 | 9.596531e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 3621.367722 | False |
| 54 | PT08.S1(CO) | RH | -2.408601 | 3.207921e-01 | -3.905366 | -3.341102 | -3.0479 | -2.405058 | 1.403139e-01 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 14436.744366 | -6.892502 | 1.344453e-09 | 7 | 1223 | -3.435708 | -2.863906 | -2.568030 | 6731.522102 | False |
| 55 | PT08.S1(CO) | AH | -2.623957 | 2.281110e-01 | -3.905366 | -3.341102 | -3.0479 | -2.623650 | 8.820710e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 14355.945648 | -3.225738 | 1.854235e-02 | 11 | 1219 | -3.435726 | -2.863914 | -2.568034 | -4136.792988 | False |
| 56 | NMHC(GT) | C6H6(GT) | -2.747914 | 1.828206e-01 | -3.905366 | -3.341102 | -3.0479 | -2.750385 | 6.571776e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 13538.402460 | -2.011819 | 2.814151e-01 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 5447.064674 | False |
| 57 | NMHC(GT) | PT08.S2(NMHC) | -2.998420 | 1.105816e-01 | -3.905366 | -3.341102 | -3.0479 | -3.000364 | 3.486044e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 13661.991740 | -2.291741 | 1.746456e-01 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 13997.932527 | False |
| 58 | NMHC(GT) | NOx(GT) | -2.721547 | 1.919250e-01 | -3.905366 | -3.341102 | -3.0479 | -2.727529 | 6.939446e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 13563.210251 | -2.344901 | 1.578951e-01 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 11225.806048 | False |
| 59 | NMHC(GT) | NO2(GT) | -2.901441 | 1.355297e-01 | -3.905366 | -3.341102 | -3.0479 | -2.906680 | 4.457675e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 13939.825696 | -2.661601 | 8.092192e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 9159.217834 | False |
| 60 | NMHC(GT) | PT08.S4(NO2) | -3.256351 | 6.099861e-02 | -3.905366 | -3.341102 | -3.0479 | -3.258184 | 1.685429e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 13679.668663 | -1.825432 | 3.679105e-01 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 14477.161825 | False |
| 61 | NMHC(GT) | PT08.S5(O3) | -2.993166 | 1.118388e-01 | -3.905366 | -3.341102 | -3.0479 | -2.996835 | 3.519103e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 13832.450803 | -2.420251 | 1.361022e-01 | 22 | 1208 | -3.435775 | -2.863936 | -2.568045 | 15036.689963 | False |
| 62 | C6H6(GT) | NOx(GT) | -2.905165 | 1.345027e-01 | -3.905366 | -3.341102 | -3.0479 | -2.904350 | 4.484453e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 5414.277645 | -3.332428 | 1.348973e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 11133.347630 | False |
| 63 | C6H6(GT) | NO2(GT) | -3.331956 | 5.049018e-02 | -3.905366 | -3.341102 | -3.0479 | -3.327442 | 1.369550e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 5836.269763 | -3.697865 | 4.143279e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 9142.759318 | False |
| 64 | C6H6(GT) | RH | -2.869318 | 1.446224e-01 | -3.905366 | -3.341102 | -3.0479 | -2.866057 | 4.943990e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 6235.586568 | -8.067092 | 1.585559e-12 | 1 | 1229 | -3.435682 | -2.863895 | -2.568024 | 6809.000977 | False |
| 65 | PT08.S2(NMHC) | NOx(GT) | -3.291918 | 5.585328e-02 | -3.905366 | -3.341102 | -3.0479 | -3.290060 | 1.532866e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 13845.204011 | -3.858865 | 2.359326e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 11110.083792 | False |
| 66 | PT08.S2(NMHC) | PT08.S3(NOx) | -3.122902 | 8.379492e-02 | -3.905366 | -3.341102 | -3.0479 | -3.132406 | 2.424611e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 13234.038489 | -3.589283 | 5.969255e-03 | 22 | 1208 | -3.435775 | -2.863936 | -2.568045 | 13027.504182 | False |
| 67 | PT08.S2(NMHC) | NO2(GT) | -3.297133 | 5.512949e-02 | -3.905366 | -3.341102 | -3.0479 | -3.291381 | 1.526817e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 14176.925809 | -3.827510 | 2.638065e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 9019.399241 | False |
| 68 | PT08.S2(NMHC) | RH | -2.967475 | 1.181406e-01 | -3.905366 | -3.341102 | -3.0479 | -2.964236 | 3.837195e-02 | 22 | 1208 | -3.435775 | -2.863936 | -2.568045 | 14590.927742 | -8.016536 | 2.131448e-12 | 1 | 1229 | -3.435682 | -2.863895 | -2.568024 | 6791.143865 | False |
| 69 | NOx(GT) | PT08.S4(NO2) | -3.705533 | 1.802743e-02 | -3.905366 | -3.341102 | -3.0479 | -3.699451 | 4.120881e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 11191.323799 | -2.487027 | 1.186628e-01 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 14377.141267 | False |
| 70 | NO2(GT) | PT08.S4(NO2) | -3.467230 | 3.541309e-02 | -3.905366 | -3.341102 | -3.0479 | -3.459714 | 9.084054e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 9148.729213 | -2.226344 | 1.968235e-01 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 14750.162495 | False |
| 71 | PT08.S4(NO2) | PT08.S5(O3) | -3.138694 | 8.079249e-02 | -3.905366 | -3.341102 | -3.0479 | -3.134582 | 2.409758e-02 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 13603.928448 | -5.111569 | 1.328716e-05 | 9 | 1221 | -3.435717 | -2.863910 | -2.568032 | 14147.451420 | False |
| 72 | PT08.S4(NO2) | T | -2.952195 | 1.220106e-01 | -3.905366 | -3.341102 | -3.0479 | -2.945505 | 4.030654e-02 | 22 | 1208 | -3.435775 | -2.863936 | -2.568045 | 15132.492318 | -3.592002 | 5.915792e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 3811.074789 | False |
| 73 | PT08.S4(NO2) | RH | -2.464276 | 2.950477e-01 | -3.905366 | -3.341102 | -3.0479 | -2.457872 | 1.260639e-01 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 15067.946106 | -6.943893 | 1.008714e-09 | 7 | 1223 | -3.435708 | -2.863906 | -2.568030 | 6755.612209 | False |
| 74 | PT08.S4(NO2) | AH | -3.498969 | 3.248692e-02 | -3.905366 | -3.341102 | -3.0479 | -3.498135 | 8.035128e-03 | 23 | 1207 | -3.435779 | -2.863938 | -2.568046 | 14986.157863 | -2.481300 | 1.200908e-01 | 22 | 1208 | -3.435775 | -2.863936 | -2.568045 | -3513.593070 | False |
| 75 | PT08.S5(O3) | T | -3.134391 | 8.160212e-02 | -3.905366 | -3.341102 | -3.0479 | -3.130477 | 2.437844e-02 | 22 | 1208 | -3.435775 | -2.863936 | -2.568045 | 15425.880947 | -3.612686 | 5.523085e-03 | 17 | 1213 | -3.435752 | -2.863926 | -2.568040 | 3593.435133 | False |
| 76 | PT08.S5(O3) | RH | -2.880129 | 1.415153e-01 | -3.905366 | -3.341102 | -3.0479 | -2.875690 | 4.824878e-02 | 22 | 1208 | -3.435775 | -2.863936 | -2.568045 | 15385.318515 | -6.892642 | 1.343400e-09 | 7 | 1223 | -3.435708 | -2.863906 | -2.568030 | 6761.987080 | False |
| 77 | T | RH | -1.903507 | 5.782269e-01 | -3.905366 | -3.341102 | -3.0479 | -1.894857 | 3.344844e-01 | 18 | 1212 | -3.435757 | -2.863928 | -2.568041 | 1866.712556 | -4.352790 | 3.594512e-04 | 9 | 1221 | -3.435717 | -2.863910 | -2.568032 | 5726.019311 | False |
Multivariate Cointegration Aanlysis
cointegration for deseasonalized time series
- NOTE: Johansen cointegration test using trace test statistic with 5% significance level
- H0: the number of cointegration vectors < r
- HA: the number of cointegration vectors = r
multivariate rolling variables(window=24): 'PT08.S1(CO)', 'PT08.S2(NMHC)', 'PT08.S3(NOx)', 'PT08.S4(NO2)', 'PT08.S5(O3)'
- cointegration rank: 4
| full rank | test statistic | critical value | reject | |
|---|---|---|---|---|
| johansen cointegration rank | ||||
| 1 | 5 | 129.988005 | 69.8189 | False |
| 2 | 5 | 70.042493 | 47.8545 | False |
| 3 | 5 | 37.747946 | 29.7961 | False |
| 4 | 5 | 14.346635 | 15.4943 | True |
multivariate rolling variables(window=24): 'PT08.S1(CO)', 'PT08.S2(NMHC)', 'PT08.S3(NOx)', 'PT08.S4(NO2)', 'PT08.S5(O3)', 'CO(GT)', 'NMHC(GT)', 'C6H6(GT)', 'NOx(GT)', 'NO2(GT)'
- cointegration rank: 7
| full rank | test statistic | critical value | reject | |
|---|---|---|---|---|
| johansen cointegration rank | ||||
| 1 | 10 | 349.008827 | 239.2468 | False |
| 2 | 10 | 249.096809 | 197.3772 | False |
| 3 | 10 | 188.240041 | 159.529 | False |
| 4 | 10 | 142.466241 | 125.6185 | False |
| 5 | 10 | 100.865666 | 95.7542 | False |
| 6 | 10 | 72.549839 | 69.8189 | False |
| 7 | 10 | 45.679593 | 47.8545 | True |
multivariate rolling variables(window=24): 'PT08.S1(CO)', 'PT08.S2(NMHC)', 'PT08.S3(NOx)', 'PT08.S4(NO2)', 'PT08.S5(O3)', 'T', 'RH', 'AH'
- cointegration rank: 7
| full rank | test statistic | critical value | reject | |
|---|---|---|---|---|
| johansen cointegration rank | ||||
| 1 | 8 | 250.292898 | 159.529 | False |
| 2 | 8 | 174.561089 | 125.6185 | False |
| 3 | 8 | 124.917753 | 95.7542 | False |
| 4 | 8 | 80.970664 | 69.8189 | False |
| 5 | 8 | 52.055663 | 47.8545 | False |
| 6 | 8 | 30.953717 | 29.7961 | False |
| 7 | 8 | 13.82569 | 15.4943 | True |
multivariate rolling variables(window=24): 'CO(GT)', 'NMHC(GT)', 'C6H6(GT)', 'NOx(GT)', 'NO2(GT)', 'T', 'RH', 'AH'
- cointegration rank: 3
| full rank | test statistic | critical value | reject | |
|---|---|---|---|---|
| johansen cointegration rank | ||||
| 1 | 8 | 184.007205 | 159.529 | False |
| 2 | 8 | 130.236397 | 125.6185 | False |
| 3 | 8 | 91.142881 | 95.7542 | True |
Cross-Covariance Analysis
'zero-lag' & 'zero-dffierence' cross covariance
: cross covariance is not steady covariance.



Univariate Time Series Characteristic Analysis
Regime Analysis

| CO(GT) | PT08.S1(CO) | NMHC(GT) | C6H6(GT) | PT08.S2(NMHC) | NOx(GT) | PT08.S3(NOx) | NO2(GT) | PT08.S4(NO2) | PT08.S5(O3) | T | RH | AH | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Evaluation | Dep. Variable: | y | y | y | y | y | y | y | y | y | y | y | y | y |
| Model: | MarkovAutoregression | MarkovAutoregression | MarkovAutoregression | MarkovAutoregression | MarkovAutoregression | MarkovAutoregression | MarkovAutoregression | MarkovAutoregression | MarkovAutoregression | MarkovAutoregression | MarkovAutoregression | MarkovAutoregression | MarkovAutoregression | |
| Date: | Sun, 06 Aug 2023 | Sun, 06 Aug 2023 | Sun, 06 Aug 2023 | Sun, 06 Aug 2023 | Sun, 06 Aug 2023 | Sun, 06 Aug 2023 | Sun, 06 Aug 2023 | Sun, 06 Aug 2023 | Sun, 06 Aug 2023 | Sun, 06 Aug 2023 | Sun, 06 Aug 2023 | Sun, 06 Aug 2023 | Sun, 06 Aug 2023 | |
| Time: | 16:10:28 | 16:10:28 | 16:10:28 | 16:10:28 | 16:10:28 | 16:10:28 | 16:10:28 | 16:10:28 | 16:10:28 | 16:10:28 | 16:10:28 | 16:10:28 | 16:10:28 | |
| Sample: | 03-10-2004 | 03-10-2004 | 03-10-2004 | 03-10-2004 | 03-10-2004 | 03-10-2004 | 03-10-2004 | 03-10-2004 | 03-10-2004 | 03-10-2004 | 03-10-2004 | 03-10-2004 | 03-10-2004 | |
| - 05-01-2004 | - 05-01-2004 | - 05-01-2004 | - 05-01-2004 | - 05-01-2004 | - 05-01-2004 | - 05-01-2004 | - 05-01-2004 | - 05-01-2004 | - 05-01-2004 | - 05-01-2004 | - 05-01-2004 | - 05-01-2004 | ||
| Covariance Type: | approx | approx | approx | approx | approx | approx | approx | approx | approx | approx | approx | approx | approx | |
| No. Observations: | 1226 | 1226 | 1226 | 1226 | 1226 | 1226 | 1226 | 1226 | 1226 | 1226 | 1226 | 1226 | 1226 | |
| Log Likelihood | -930.211 | -7365.290 | -6959.116 | -3012.664 | -7429.182 | -5944.079 | -7262.418 | -4694.032 | -7692.651 | -7811.766 | -1582.347 | -3275.844 | 2496.972 | |
| AIC | 1876.421 | 14746.580 | 13934.233 | 6041.327 | 14874.364 | 11904.159 | 14540.835 | 9404.065 | 15401.303 | 15639.533 | 3180.694 | 6567.687 | -4977.944 | |
| BIC | 1917.313 | 14787.472 | 13975.125 | 6082.219 | 14915.256 | 11945.051 | 14581.727 | 9444.957 | 15442.195 | 15680.425 | 3221.586 | 6608.579 | -4937.052 | |
| HQIC | 1891.809 | 14761.968 | 13949.620 | 6056.715 | 14889.751 | 11919.546 | 14556.223 | 9419.452 | 15416.690 | 15654.920 | 3196.081 | 6583.075 | -4962.557 | |
| Parameter | p[0->0] | 0.880076 | 0.428455 | 0.876458 | 0.796967 | 0.426028 | 0.893698 | 0.390147 | 0.981973 | 0.465543 | 0.415082 | 0.901006 | 0.960652 | 0.920954 |
| p[1->0] | 0.214701 | 0.362537 | 0.170316 | 0.218055 | 0.361342 | 0.161763 | 0.352856 | 0.003532 | 0.37483 | 0.358166 | 0.13585 | 0.198772 | 0.179942 | |
| sigma2[0] | 0.084451 | 9675.113302 | 948.035153 | 1.535768 | 10737.746091 | 317.025499 | 8172.310031 | 13.687741 | 16503.04379 | 20042.703687 | 0.290376 | 7.299175 | 0.000353 | |
| sigma2[1] | 0.869331 | 9674.575384 | 18974.172769 | 20.595025 | 10737.497575 | 2616.902935 | 8184.612921 | 187.285781 | 16503.30315 | 20042.829075 | 1.765689 | 60.494245 | 0.004115 | |
| ar.L1 | 0.183671 | 0.092349 | 0.188038 | 0.128529 | 0.128017 | 0.154356 | 0.181609 | 0.120595 | 0.064329 | 0.147155 | 0.276113 | 0.240807 | 0.269414 | |
| ar.L2 | -0.173284 | -0.122451 | -0.20014 | -0.110626 | -0.127944 | -0.088726 | -0.100084 | -0.109547 | -0.121187 | -0.112021 | 0.041176 | -0.01256 | -0.083374 | |
| ar.L3 | -0.059278 | -0.032816 | -0.08476 | -0.08827 | -0.065946 | -0.024228 | -0.100173 | -0.070878 | -0.068672 | -0.062568 | 0.017834 | 0.00195 | 0.015232 | |
| ar.L4 | -0.017732 | -0.072015 | -0.016172 | -0.000896 | -0.062958 | -0.012737 | -0.101756 | -0.066468 | -0.041504 | -0.086943 | -0.017472 | 0.027337 | 0.015456 | |
| duration[0] | 8.33864 | 1.749642 | 8.094415 | 4.925317 | 1.742246 | 9.407152 | 1.63974 | 55.472493 | 1.871059 | 1.709641 | 10.10161 | 25.413964 | 12.650861 | |
| duration[1] | 4.657631 | 2.758343 | 5.871432 | 4.586005 | 2.767463 | 6.181893 | 2.834013 | 283.100248 | 2.667875 | 2.792 | 7.361083 | 5.030878 | 5.557336 |
Volatility Analysis

| CO(GT) | PT08.S1(CO) | NMHC(GT) | C6H6(GT) | PT08.S2(NMHC) | NOx(GT) | PT08.S3(NOx) | NO2(GT) | PT08.S4(NO2) | PT08.S5(O3) | T | RH | AH | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | |||||||||||||
| Dep. Variable: | CO(GT) | PT08.S1(CO) | NMHC(GT) | C6H6(GT) | PT08.S2(NMHC) | NOx(GT) | PT08.S3(NOx) | NO2(GT) | PT08.S4(NO2) | PT08.S5(O3) | T | RH | AH |
| Mean Model: | AR | AR | AR | AR | AR | AR | AR | AR | AR | AR | AR | AR | AR |
| Vol Model: | GJR-GARCH | GJR-GARCH | GJR-GARCH | GJR-GARCH | GJR-GARCH | GJR-GARCH | GJR-GARCH | GJR-GARCH | GJR-GARCH | GJR-GARCH | GJR-GARCH | GJR-GARCH | GJR-GARCH |
| Distribution: | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal |
| Method: | Maximum Likelihood | Maximum Likelihood | Maximum Likelihood | Maximum Likelihood | Maximum Likelihood | Maximum Likelihood | Maximum Likelihood | Maximum Likelihood | Maximum Likelihood | Maximum Likelihood | Maximum Likelihood | Maximum Likelihood | Maximum Likelihood |
| Date: | Sun, Aug 06 2023 | Sun, Aug 06 2023 | Sun, Aug 06 2023 | Sun, Aug 06 2023 | Sun, Aug 06 2023 | Sun, Aug 06 2023 | Sun, Aug 06 2023 | Sun, Aug 06 2023 | Sun, Aug 06 2023 | Sun, Aug 06 2023 | Sun, Aug 06 2023 | Sun, Aug 06 2023 | Sun, Aug 06 2023 |
| Time: | 16:49:42 | 16:49:43 | 16:49:43 | 16:49:43 | 16:49:44 | 16:49:44 | 16:49:44 | 16:49:45 | 16:49:45 | 16:49:45 | 16:49:46 | 16:49:46 | 16:49:46 |
| R-squared: | 0.026 | 0.027 | 0.012 | 0.023 | 0.039 | 0.016 | 0.069 | 0.005 | 0.018 | 0.044 | 0.080 | 0.052 | 0.047 |
| Adj. R-squared: | 0.023 | 0.023 | 0.009 | 0.020 | 0.036 | 0.013 | 0.066 | 0.002 | 0.015 | 0.041 | 0.077 | 0.049 | 0.044 |
| Log-Likelihood: | -1044.95 | -7343.31 | -7150.36 | -3085.62 | -7390.33 | -6020.84 | -7232.22 | -4732.09 | -7627.62 | -7795.19 | -1625.91 | -3322.75 | 2365.46 |
| AIC: | 2111.90 | 14708.6 | 14322.7 | 6193.24 | 14802.7 | 12063.7 | 14486.4 | 9486.19 | 15277.2 | 15612.4 | 3273.81 | 6667.49 | -4708.93 |
| BIC: | 2168.13 | 14764.8 | 14378.9 | 6249.46 | 14858.9 | 12119.9 | 14542.7 | 9542.41 | 15333.5 | 15668.6 | 3330.04 | 6723.72 | -4652.70 |
| No. Observations: | 1226 | 1226 | 1226 | 1226 | 1226 | 1226 | 1226 | 1226 | 1226 | 1226 | 1226 | 1226 | 1226 |
| Df Residuals: | 1221 | 1221 | 1221 | 1221 | 1221 | 1221 | 1221 | 1221 | 1221 | 1221 | 1221 | 1221 | 1221 |
| Df Model: | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| coef | std err | t | P>|t| | 95.0% Conf. Int. | |||
|---|---|---|---|---|---|---|---|
| variable | characteristics type | ||||||
| CO(GT) | Mean | Const | -0.0132 | 1.515e-02 | -0.869 | 0.385 | [-4.286e-02,1.653e-02] |
| CO(GT)[1] | 0.2175 | 4.527e-02 | 4.803 | 1.561e-06 | [ 0.129, 0.306] | ||
| CO(GT)[2] | -0.1925 | 4.592e-02 | -4.192 | 2.766e-05 | [ -0.283, -0.102] | ||
| CO(GT)[3] | -0.0127 | 3.291e-02 | -0.385 | 0.700 | [-7.719e-02,5.184e-02] | ||
| CO(GT)[4] | -0.0229 | 3.475e-02 | -0.660 | 0.509 | [-9.105e-02,4.517e-02] | ||
| PT08.S1(CO) | Mean | Const | 0.9198 | 3.047 | 0.302 | 0.763 | [ -5.053, 6.892] |
| PT08...CO)[1] | 0.1306 | 3.456e-02 | 3.779 | 1.574e-04 | [6.286e-02, 0.198] | ||
| PT08...CO)[2] | -0.1122 | 3.132e-02 | -3.583 | 3.395e-04 | [ -0.174,-5.084e-02] | ||
| PT08...CO)[3] | -0.0277 | 3.272e-02 | -0.846 | 0.398 | [-9.181e-02,3.646e-02] | ||
| PT08...CO)[4] | -0.0566 | 2.492e-02 | -2.269 | 2.324e-02 | [ -0.105,-7.714e-03] | ||
| NMHC(GT) | Mean | Const | -1.2142 | 2.567 | -0.473 | 0.636 | [ -6.245, 3.816] |
| NMHC(GT)[1] | 0.2326 | 6.858e-02 | 3.392 | 6.940e-04 | [9.820e-02, 0.367] | ||
| NMHC(GT)[2] | -0.2214 | 5.068e-02 | -4.368 | 1.252e-05 | [ -0.321, -0.122] | ||
| NMHC(GT)[3] | -5.9363e-03 | 4.593e-02 | -0.129 | 0.897 | [-9.596e-02,8.409e-02] | ||
| NMHC(GT)[4] | -0.0450 | 3.409e-02 | -1.320 | 0.187 | [ -0.112,2.183e-02] | ||
| C6H6(GT) | Mean | Const | 0.0156 | 7.820e-02 | 0.199 | 0.842 | [ -0.138, 0.169] |
| C6H6(GT)[1] | 0.2141 | 4.681e-02 | 4.575 | 4.771e-06 | [ 0.122, 0.306] | ||
| C6H6(GT)[2] | -0.1480 | 3.487e-02 | -4.245 | 2.187e-05 | [ -0.216,-7.968e-02] | ||
| C6H6(GT)[3] | -0.0592 | 2.376e-02 | -2.493 | 1.265e-02 | [ -0.106,-1.267e-02] | ||
| C6H6(GT)[4] | -3.6311e-03 | 2.243e-02 | -0.162 | 0.871 | [-4.760e-02,4.034e-02] | ||
| PT08.S2(NMHC) | Mean | Const | 1.5523 | 2.763 | 0.562 | 0.574 | [ -3.864, 6.968] |
| PT08...HC)[1] | 0.1625 | 3.585e-02 | 4.532 | 5.840e-06 | [9.222e-02, 0.233] | ||
| PT08...HC)[2] | -0.1342 | 2.936e-02 | -4.571 | 4.847e-06 | [ -0.192,-7.668e-02] | ||
| PT08...HC)[3] | -0.0695 | 2.577e-02 | -2.696 | 7.016e-03 | [ -0.120,-1.897e-02] | ||
| PT08...HC)[4] | -0.0288 | 2.605e-02 | -1.105 | 0.269 | [-7.984e-02,2.227e-02] | ||
| NOx(GT) | Mean | Const | -0.4985 | 1.027 | -0.485 | 0.627 | [ -2.512, 1.514] |
| NOx(GT)[1] | 0.1356 | 4.180e-02 | 3.243 | 1.183e-03 | [5.364e-02, 0.218] | ||
| NOx(GT)[2] | -0.1055 | 3.981e-02 | -2.649 | 8.073e-03 | [ -0.183,-2.743e-02] | ||
| NOx(GT)[3] | -0.0620 | 3.784e-02 | -1.638 | 0.101 | [ -0.136,1.217e-02] | ||
| NOx(GT)[4] | -0.0337 | 3.955e-02 | -0.852 | 0.394 | [ -0.111,4.382e-02] | ||
| PT08.S3(NOx) | Mean | Const | -2.9614 | 2.683 | -1.104 | 0.270 | [ -8.220, 2.298] |
| PT08...Ox)[1] | 0.1586 | 3.192e-02 | 4.968 | 6.767e-07 | [9.601e-02, 0.221] | ||
| PT08...Ox)[2] | -0.0970 | 3.147e-02 | -3.084 | 2.044e-03 | [ -0.159,-3.536e-02] | ||
| PT08...Ox)[3] | -0.1078 | 3.045e-02 | -3.542 | 3.973e-04 | [ -0.168,-4.817e-02] | ||
| PT08...Ox)[4] | -0.1017 | 3.004e-02 | -3.385 | 7.110e-04 | [ -0.161,-4.282e-02] | ||
| NO2(GT) | Mean | Const | -0.1453 | 0.272 | -0.535 | 0.593 | [ -0.678, 0.387] |
| NO2(GT)[1] | 0.0610 | 3.600e-02 | 1.696 | 8.998e-02 | [-9.521e-03, 0.132] | ||
| NO2(GT)[2] | -0.0943 | 3.248e-02 | -2.904 | 3.686e-03 | [ -0.158,-3.066e-02] | ||
| NO2(GT)[3] | -0.0505 | 3.376e-02 | -1.496 | 0.135 | [ -0.117,1.568e-02] | ||
| NO2(GT)[4] | -0.0574 | 3.011e-02 | -1.907 | 5.650e-02 | [ -0.116,1.591e-03] | ||
| PT08.S4(NO2) | Mean | Const | 1.0498 | 3.409 | 0.308 | 0.758 | [ -5.631, 7.731] |
| PT08...O2)[1] | 0.1493 | 4.324e-02 | 3.452 | 5.572e-04 | [6.451e-02, 0.234] | ||
| PT08...O2)[2] | -0.1248 | 3.086e-02 | -4.042 | 5.299e-05 | [ -0.185,-6.426e-02] | ||
| PT08...O2)[3] | -0.0599 | 2.494e-02 | -2.400 | 1.638e-02 | [ -0.109,-1.098e-02] | ||
| PT08...O2)[4] | -8.6271e-03 | 2.357e-02 | -0.366 | 0.714 | [-5.482e-02,3.757e-02] | ||
| PT08.S5(O3) | Mean | Const | 0.3936 | 12.089 | 3.256e-02 | 0.974 | [-23.301, 24.089] |
| PT08...O3)[1] | 0.1976 | 4.808e-02 | 4.110 | 3.955e-05 | [ 0.103, 0.292] | ||
| PT08...O3)[2] | -0.1282 | 3.233e-02 | -3.966 | 7.307e-05 | [ -0.192,-6.487e-02] | ||
| PT08...O3)[3] | -0.0413 | 6.376e-02 | -0.648 | 0.517 | [ -0.166,8.368e-02] | ||
| PT08...O3)[4] | -0.0695 | 4.573e-02 | -1.521 | 0.128 | [ -0.159,2.009e-02] | ||
| T | Mean | Const | -2.9876e-04 | 2.519e-02 | -1.186e-02 | 0.991 | [-4.967e-02,4.908e-02] |
| T[1] | 0.2962 | 4.306e-02 | 6.880 | 5.997e-12 | [ 0.212, 0.381] | ||
| T[2] | -0.0314 | 4.427e-02 | -0.709 | 0.478 | [ -0.118,5.535e-02] | ||
| T[3] | 0.0495 | 3.918e-02 | 1.264 | 0.206 | [-2.726e-02, 0.126] | ||
| T[4] | -0.0396 | 3.018e-02 | -1.313 | 0.189 | [-9.876e-02,1.953e-02] | ||
| RH | Mean | Const | 0.0186 | 0.103 | 0.180 | 0.857 | [ -0.184, 0.221] |
| RH[1] | 0.2422 | 3.983e-02 | 6.081 | 1.192e-09 | [ 0.164, 0.320] | ||
| RH[2] | -0.0514 | 5.091e-02 | -1.010 | 0.313 | [ -0.151,4.837e-02] | ||
| RH[3] | 0.0544 | 4.568e-02 | 1.191 | 0.234 | [-3.514e-02, 0.144] | ||
| RH[4] | -0.0181 | 3.271e-02 | -0.555 | 0.579 | [-8.226e-02,4.596e-02] | ||
| AH | Mean | Const | -3.8408e-04 | 1.721e-03 | -0.223 | 0.823 | [-3.757e-03,2.989e-03] |
| AH[1] | 0.2611 | 4.776e-02 | 5.467 | 4.584e-08 | [ 0.167, 0.355] | ||
| AH[2] | -0.1206 | 4.646e-02 | -2.596 | 9.430e-03 | [ -0.212,-2.955e-02] | ||
| AH[3] | 0.0233 | 4.310e-02 | 0.541 | 0.588 | [-6.116e-02, 0.108] | ||
| AH[4] | 0.0146 | 4.965e-02 | 0.294 | 0.769 | [-8.272e-02, 0.112] | ||
| CO(GT) | Volatility | omega | 0.0177 | 3.048e-02 | 0.581 | 0.561 | [-4.203e-02,7.746e-02] |
| alpha[1] | 0.1239 | 7.244e-02 | 1.711 | 8.716e-02 | [-1.806e-02, 0.266] | ||
| alpha[2] | 0.0000 | 0.108 | 0.000 | 1.000 | [ -0.212, 0.212] | ||
| gamma[1] | -0.0942 | 0.147 | -0.640 | 0.522 | [ -0.383, 0.194] | ||
| beta[1] | 0.5241 | 0.708 | 0.740 | 0.459 | [ -0.864, 1.913] | ||
| beta[2] | 0.3452 | 0.561 | 0.615 | 0.539 | [ -0.755, 1.446] | ||
| PT08.S1(CO) | Volatility | omega | 4837.4131 | 3873.570 | 1.249 | 0.212 | [-2.755e+03,1.243e+04] |
| alpha[1] | 0.2050 | 0.139 | 1.473 | 0.141 | [-6.786e-02, 0.478] | ||
| alpha[2] | 6.2117e-12 | 4.262e-02 | 1.458e-10 | 1.000 | [-8.353e-02,8.353e-02] | ||
| gamma[1] | -0.1817 | 0.100 | -1.815 | 6.956e-02 | [ -0.378,1.454e-02] | ||
| beta[1] | 0.3744 | 0.482 | 0.777 | 0.437 | [ -0.571, 1.319] | ||
| beta[2] | 3.5600e-13 | 0.898 | 3.965e-13 | 1.000 | [ -1.760, 1.760] | ||
| NMHC(GT) | Volatility | omega | 3565.6077 | 1569.228 | 2.272 | 2.307e-02 | [4.900e+02,6.641e+03] |
| alpha[1] | 0.7221 | 0.226 | 3.198 | 1.382e-03 | [ 0.280, 1.165] | ||
| alpha[2] | 0.1757 | 0.283 | 0.621 | 0.535 | [ -0.379, 0.730] | ||
| gamma[1] | -0.5062 | 0.203 | -2.492 | 1.271e-02 | [ -0.904, -0.108] | ||
| beta[1] | 0.0000 | 0.499 | 0.000 | 1.000 | [ -0.977, 0.977] | ||
| beta[2] | 0.0639 | 0.140 | 0.457 | 0.648 | [ -0.210, 0.338] | ||
| C6H6(GT) | Volatility | omega | 4.5609 | 1.028 | 4.437 | 9.105e-06 | [ 2.546, 6.575] |
| alpha[1] | 0.8608 | 0.245 | 3.517 | 4.360e-04 | [ 0.381, 1.340] | ||
| alpha[2] | 0.0000 | 9.654e-02 | 0.000 | 1.000 | [ -0.189, 0.189] | ||
| gamma[1] | -0.5263 | 0.177 | -2.971 | 2.969e-03 | [ -0.874, -0.179] | ||
| beta[1] | 0.1196 | 0.133 | 0.897 | 0.370 | [ -0.142, 0.381] | ||
| beta[2] | 0.0000 | 5.046e-02 | 0.000 | 1.000 | [-9.890e-02,9.890e-02] | ||
| PT08.S2(NMHC) | Volatility | omega | 5896.4065 | 1320.354 | 4.466 | 7.978e-06 | [3.309e+03,8.484e+03] |
| alpha[1] | 0.3639 | 8.802e-02 | 4.134 | 3.563e-05 | [ 0.191, 0.536] | ||
| alpha[2] | 8.9518e-13 | 6.096e-02 | 1.469e-11 | 1.000 | [ -0.119, 0.119] | ||
| gamma[1] | -0.1607 | 6.589e-02 | -2.440 | 1.471e-02 | [ -0.290,-3.160e-02] | ||
| beta[1] | 0.1893 | 0.201 | 0.940 | 0.347 | [ -0.205, 0.584] | ||
| beta[2] | 2.3487e-12 | 0.110 | 2.133e-11 | 1.000 | [ -0.216, 0.216] | ||
| NOx(GT) | Volatility | omega | 25.1199 | 33.212 | 0.756 | 0.449 | [-39.975, 90.214] |
| alpha[1] | 0.0989 | 4.605e-02 | 2.148 | 3.175e-02 | [8.637e-03, 0.189] | ||
| alpha[2] | 1.2340e-13 | 5.605e-02 | 2.202e-12 | 1.000 | [ -0.110, 0.110] | ||
| gamma[1] | -0.0134 | 0.100 | -0.133 | 0.894 | [ -0.210, 0.183] | ||
| beta[1] | 0.1890 | 0.129 | 1.460 | 0.144 | [-6.476e-02, 0.443] | ||
| beta[2] | 0.7009 | 0.116 | 6.016 | 1.783e-09 | [ 0.473, 0.929] | ||
| PT08.S3(NOx) | Volatility | omega | 2454.0247 | 1954.886 | 1.255 | 0.209 | [-1.377e+03,6.286e+03] |
| alpha[1] | 0.2455 | 7.112e-02 | 3.451 | 5.583e-04 | [ 0.106, 0.385] | ||
| alpha[2] | 0.0576 | 9.412e-02 | 0.612 | 0.540 | [ -0.127, 0.242] | ||
| gamma[1] | -0.1941 | 7.508e-02 | -2.585 | 9.742e-03 | [ -0.341,-4.692e-02] | ||
| beta[1] | 0.2029 | 0.316 | 0.642 | 0.521 | [ -0.417, 0.822] | ||
| beta[2] | 0.2946 | 0.113 | 2.617 | 8.859e-03 | [7.401e-02, 0.515] | ||
| NO2(GT) | Volatility | omega | 1.3273 | 0.983 | 1.350 | 0.177 | [ -0.599, 3.254] |
| alpha[1] | 0.1663 | 4.701e-02 | 3.538 | 4.026e-04 | [7.420e-02, 0.258] | ||
| alpha[2] | 7.1748e-09 | 0.116 | 6.185e-08 | 1.000 | [ -0.227, 0.227] | ||
| gamma[1] | -0.0188 | 3.703e-02 | -0.509 | 0.611 | [-9.141e-02,5.373e-02] | ||
| beta[1] | 0.4981 | 0.620 | 0.804 | 0.422 | [ -0.717, 1.713] | ||
| beta[2] | 0.3450 | 0.539 | 0.640 | 0.522 | [ -0.712, 1.402] | ||
| PT08.S4(NO2) | Volatility | omega | 9219.9300 | 1976.741 | 4.664 | 3.098e-06 | [5.346e+03,1.309e+04] |
| alpha[1] | 0.5076 | 0.135 | 3.751 | 1.761e-04 | [ 0.242, 0.773] | ||
| alpha[2] | 7.5450e-12 | 6.056e-02 | 1.246e-10 | 1.000 | [ -0.119, 0.119] | ||
| gamma[1] | -0.2908 | 0.131 | -2.216 | 2.669e-02 | [ -0.548,-3.361e-02] | ||
| beta[1] | 0.1184 | 0.169 | 0.702 | 0.483 | [ -0.212, 0.449] | ||
| beta[2] | 2.2014e-12 | 6.307e-02 | 3.490e-11 | 1.000 | [ -0.124, 0.124] | ||
| PT08.S5(O3) | Volatility | omega | 1.0021e+04 | 3.009e+04 | 0.333 | 0.739 | [-4.896e+04,6.900e+04] |
| alpha[1] | 0.2182 | 0.132 | 1.652 | 9.844e-02 | [-4.060e-02, 0.477] | ||
| alpha[2] | 1.6768e-13 | 0.692 | 2.422e-13 | 1.000 | [ -1.357, 1.357] | ||
| gamma[1] | -0.1001 | 5.897e-02 | -1.697 | 8.977e-02 | [ -0.216,1.553e-02] | ||
| beta[1] | 0.3364 | 0.361 | 0.933 | 0.351 | [ -0.370, 1.043] | ||
| beta[2] | 6.7762e-13 | 1.940 | 3.492e-13 | 1.000 | [ -3.803, 3.803] | ||
| T | Volatility | omega | 0.3046 | 7.903e-02 | 3.854 | 1.160e-04 | [ 0.150, 0.460] |
| alpha[1] | 0.2030 | 6.964e-02 | 2.914 | 3.564e-03 | [6.646e-02, 0.339] | ||
| alpha[2] | 0.1384 | 7.641e-02 | 1.811 | 7.016e-02 | [-1.139e-02, 0.288] | ||
| gamma[1] | 0.1468 | 6.618e-02 | 2.218 | 2.658e-02 | [1.706e-02, 0.276] | ||
| beta[1] | 1.1683e-11 | 0.162 | 7.193e-11 | 1.000 | [ -0.318, 0.318] | ||
| beta[2] | 0.2960 | 8.868e-02 | 3.337 | 8.454e-04 | [ 0.122, 0.470] | ||
| RH | Volatility | omega | 4.5112 | 1.034 | 4.363 | 1.282e-05 | [ 2.485, 6.538] |
| alpha[1] | 0.3636 | 9.310e-02 | 3.906 | 9.398e-05 | [ 0.181, 0.546] | ||
| alpha[2] | 0.1869 | 8.597e-02 | 2.174 | 2.967e-02 | [1.844e-02, 0.355] | ||
| gamma[1] | -0.2554 | 7.789e-02 | -3.278 | 1.044e-03 | [ -0.408, -0.103] | ||
| beta[1] | 7.4700e-10 | 0.187 | 3.991e-09 | 1.000 | [ -0.367, 0.367] | ||
| beta[2] | 0.3207 | 0.140 | 2.295 | 2.172e-02 | [4.684e-02, 0.595] | ||
| AH | Volatility | omega | 4.6079e-04 | 4.063e-04 | 1.134 | 0.257 | [-3.355e-04,1.257e-03] |
| alpha[1] | 0.1009 | 3.929e-02 | 2.568 | 1.022e-02 | [2.390e-02, 0.178] | ||
| alpha[2] | 0.1004 | 0.240 | 0.418 | 0.676 | [ -0.371, 0.571] | ||
| gamma[1] | 0.1988 | 0.259 | 0.767 | 0.443 | [ -0.309, 0.707] | ||
| beta[1] | 0.1998 | 0.640 | 0.312 | 0.755 | [ -1.054, 1.453] | ||
| beta[2] | 0.1998 | 0.254 | 0.786 | 0.432 | [ -0.299, 0.698] |
Autocorrelation Analysis
ACF and PACF
Autocorrelation for differencing (D=1, d=1, m=24)
| CO(GT) | PT08.S1(CO) | NMHC(GT) | C6H6(GT) | PT08.S2(NMHC) | NOx(GT) | PT08.S3(NOx) | NO2(GT) | PT08.S4(NO2) | PT08.S5(O3) | T | RH | AH | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ref:acov | ref:pacorr | acorr | stats | p-val | determination | ref:acov | ref:pacorr | acorr | stats | p-val | determination | ref:acov | ref:pacorr | acorr | stats | p-val | determination | ref:acov | ref:pacorr | acorr | stats | p-val | determination | ref:acov | ref:pacorr | acorr | stats | p-val | determination | ref:acov | ref:pacorr | acorr | stats | p-val | determination | ref:acov | ref:pacorr | acorr | stats | p-val | determination | ref:acov | ref:pacorr | acorr | stats | p-val | determination | ref:acov | ref:pacorr | acorr | stats | p-val | determination | ref:acov | ref:pacorr | acorr | stats | p-val | determination | ref:acov | ref:pacorr | acorr | stats | p-val | determination | ref:acov | ref:pacorr | acorr | stats | p-val | determination | ref:acov | ref:pacorr | acorr | stats | p-val | determination | |
| lag0 | 0.737012 | 1.0 | 1.0 | 6.163896 | 0.013038 | True | 17538.287069 | 1.0 | 1.0 | 3.552425 | 0.059458 | False | 17726.064155 | 1.0 | 1.0 | 5.37914 | 0.020379 | True | 18.703476 | 1.0 | 1.0 | 5.1543 | 0.023189 | True | 18759.327043 | 1.0 | 1.0 | 2.006677 | 0.156608 | False | 2534.369073 | 1.0 | 1.0 | 1.938377 | 0.163845 | False | 15816.425965 | 1.0 | 1.0 | 3.180219 | 0.074535 | False | 321.789689 | 1.0 | 1.0 | 11.337678 | 0.00076 | True | 29980.260228 | 1.0 | 1.0 | 12.008053 | 0.00053 | True | 39172.865313 | 1.0 | 1.0 | 1.752227 | 0.185597 | False | 1.885387 | 1.0 | 1.0 | 43.543333 | 0.0 | True | 34.84285 | 1.0 | 1.0 | 47.325554 | 0.0 | True | 0.003098 | 1.0 | 1.0 | 48.856119 | 0.0 | True |
| lag1 | -0.052625 | -0.071462 | -0.071403 | 49.113444 | 0.0 | True | -950.683719 | -0.054251 | -0.054206 | 34.99744 | 0.0 | True | -1182.374765 | -0.066758 | -0.066703 | 52.311442 | 0.0 | True | -1.221219 | -0.065348 | -0.065294 | 38.372532 | 0.0 | True | -764.26223 | -0.040774 | -0.04074 | 26.239749 | 0.000002 | True | -101.478816 | -0.040074 | -0.040041 | 39.692093 | 0.0 | True | 811.191314 | 0.05133 | 0.051288 | 20.078824 | 0.000044 | True | -31.161659 | -0.096919 | -0.096839 | 46.773112 | 0.0 | True | -2987.84526 | -0.099743 | -0.09966 | 39.6927 | 0.0 | True | 1491.30659 | 0.038101 | 0.03807 | 25.047398 | 0.000004 | True | 0.357806 | 0.189936 | 0.189779 | 43.881213 | 0.0 | True | 6.893629 | 0.198013 | 0.197849 | 47.534531 | 0.0 | True | 0.000623 | 0.20119 | 0.201023 | 61.849397 | 0.0 | True |
| lag2 | -0.138855 | -0.194817 | -0.188402 | 63.412334 | 0.0 | True | -2827.28245 | -0.164903 | -0.161206 | 47.800784 | 0.0 | True | -3491.036302 | -0.202631 | -0.196944 | 66.33416 | 0.0 | True | -3.098965 | -0.170965 | -0.165689 | 57.948788 | 0.0 | True | -2654.774326 | -0.143654 | -0.141518 | 43.78898 | 0.0 | True | -447.667779 | -0.178825 | -0.176639 | 52.345664 | 0.0 | True | -1869.131657 | -0.121327 | -0.118177 | 49.391281 | 0.0 | True | -55.067724 | -0.182522 | -0.17113 | 54.410695 | 0.0 | True | -4534.830004 | -0.163083 | -0.151261 | 53.802565 | 0.0 | True | -5435.311141 | -0.140638 | -0.138752 | 49.1044 | 0.0 | True | 0.031506 | -0.020061 | 0.01671 | 44.657789 | 0.0 | True | 0.457898 | -0.027109 | 0.013142 | 50.920151 | 0.0 | True | -0.000321 | -0.150361 | -0.103625 | 67.276734 | 0.0 | True |
| lag3 | -0.080085 | -0.145192 | -0.108662 | 80.631299 | 0.0 | True | -1803.327384 | -0.126017 | -0.102822 | 68.527366 | 0.0 | True | -1907.454155 | -0.143758 | -0.107607 | 82.624071 | 0.0 | True | -2.378005 | -0.156556 | -0.127142 | 69.147991 | 0.0 | True | -2258.249875 | -0.13618 | -0.12038 | 62.924353 | 0.0 | True | -259.061032 | -0.122093 | -0.102219 | 82.902653 | 0.0 | True | -2460.708455 | -0.145445 | -0.155579 | 87.554728 | 0.0 | True | -25.554993 | -0.122126 | -0.079415 | 74.926652 | 0.0 | True | -3236.104442 | -0.147401 | -0.107941 | 68.072641 | 0.0 | True | -5521.178493 | -0.13271 | -0.140944 | 70.332718 | 0.0 | True | -0.047744 | -0.025758 | -0.025323 | 44.681019 | 0.0 | True | -1.842292 | -0.052407 | -0.052874 | 50.927943 | 0.0 | True | -0.000207 | -0.013689 | -0.066945 | 69.718787 | 0.0 | True |
| lag4 | -0.087846 | -0.19366 | -0.119192 | 80.631519 | 0.0 | True | -2293.486173 | -0.182928 | -0.13077 | 69.415748 | 0.0 | True | -2055.023499 | -0.192492 | -0.115932 | 82.812628 | 0.0 | True | -1.797881 | -0.159293 | -0.096126 | 69.427149 | 0.0 | True | -2357.115381 | -0.167645 | -0.12565 | 62.95946 | 0.0 | True | -402.411383 | -0.214211 | -0.158782 | 82.930497 | 0.0 | True | -2806.578919 | -0.184758 | -0.177447 | 94.364314 | 0.0 | True | -41.86616 | -0.198191 | -0.130104 | 75.699105 | 0.0 | True | -3253.071825 | -0.176101 | -0.108507 | 68.084009 | 0.0 | True | -5184.277562 | -0.148684 | -0.132344 | 72.230234 | 0.0 | True | -0.008254 | 0.005875 | -0.004378 | 51.677494 | 0.0 | True | 0.088345 | 0.024903 | 0.002536 | 58.555965 | 0.0 | True | -0.000139 | -0.04497 | -0.044887 | 71.266103 | 0.0 | True |
| lag5 | -0.000314 | -0.099489 | -0.000426 | 80.815052 | 0.0 | True | -474.625968 | -0.103023 | -0.027062 | 69.451831 | 0.0 | True | -221.00259 | -0.112609 | -0.012468 | 82.859092 | 0.0 | True | 0.283734 | -0.068866 | 0.01517 | 69.728816 | 0.0 | True | -100.920285 | -0.070205 | -0.00538 | 63.035955 | 0.0 | True | -12.142112 | -0.083395 | -0.004791 | 83.117336 | 0.0 | True | -1185.040275 | -0.108333 | -0.074925 | 95.038818 | 0.0 | True | 8.120312 | -0.065356 | 0.025235 | 75.983375 | 0.0 | True | 91.777355 | -0.089284 | 0.003061 | 68.175089 | 0.0 | True | -1549.324595 | -0.077848 | -0.039551 | 73.143856 | 0.0 | True | -0.143187 | -0.079408 | -0.075946 | 71.029727 | 0.0 | True | -2.76302 | -0.089012 | -0.079299 | 64.954242 | 0.0 | True | -0.000111 | -0.029053 | -0.035715 | 71.282056 | 0.0 | True |
| lag6 | 0.009062 | -0.09687 | 0.012295 | 80.904878 | 0.0 | True | 95.614679 | -0.088009 | 0.005452 | 69.74926 | 0.0 | True | 109.662433 | -0.109188 | 0.006187 | 82.873872 | 0.0 | True | -0.294829 | -0.100193 | -0.015763 | 70.131362 | 0.0 | True | -148.907228 | -0.084777 | -0.007938 | 63.037163 | 0.0 | True | -31.440349 | -0.120977 | -0.012406 | 83.879986 | 0.0 | True | 372.807059 | -0.047119 | 0.023571 | 97.691537 | 0.0 | True | 4.924028 | -0.074736 | 0.015302 | 76.138517 | 0.0 | True | 259.675 | -0.081631 | 0.008662 | 68.23488 | 0.0 | True | -1074.61328 | -0.093421 | -0.027433 | 73.346024 | 0.0 | True | -0.23804 | -0.102597 | -0.126255 | 91.775743 | 0.0 | True | -2.529465 | -0.04488 | -0.072596 | 83.454621 | 0.0 | True | -0.000011 | -0.001151 | -0.003625 | 72.597799 | 0.0 | True |
| lag7 | -0.006337 | -0.097095 | -0.008598 | 85.203179 | 0.0 | True | 274.397996 | -0.064377 | 0.015646 | 70.921132 | 0.0 | True | -61.821501 | -0.102148 | -0.003488 | 83.451984 | 0.0 | True | -0.340434 | -0.089945 | -0.018202 | 72.926253 | 0.0 | True | 18.706093 | -0.065399 | 0.000997 | 65.296565 | 0.0 | True | 63.494347 | -0.059731 | 0.025053 | 87.134898 | 0.0 | True | 739.020414 | -0.03617 | 0.046725 | 97.719562 | 0.0 | True | -3.636128 | -0.071159 | -0.0113 | 76.151424 | 0.0 | True | 210.307732 | -0.066235 | 0.007015 | 71.153222 | 0.0 | True | 505.293117 | -0.049076 | 0.012899 | 73.42871 | 0.0 | True | -0.24636 | -0.093128 | -0.130668 | 95.346259 | 0.0 | True | -4.299386 | -0.104102 | -0.123394 | 92.565783 | 0.0 | True | -0.000102 | -0.046511 | -0.032907 | 77.366577 | 0.0 | True |
| lag8 | -0.043817 | -0.150763 | -0.059452 | 85.275192 | 0.0 | True | -544.438301 | -0.105084 | -0.031043 | 73.170773 | 0.0 | True | -386.491353 | -0.118468 | -0.021804 | 84.929925 | 0.0 | True | -0.896656 | -0.122254 | -0.047941 | 73.410712 | 0.0 | True | -808.60289 | -0.106831 | -0.043104 | 66.005137 | 0.0 | True | -131.117528 | -0.148242 | -0.051736 | 89.322231 | 0.0 | True | 75.928346 | -0.06532 | 0.004801 | 98.053553 | 0.0 | True | -1.048378 | -0.064148 | -0.003258 | 76.166544 | 0.0 | True | -1468.672093 | -0.115216 | -0.048988 | 72.671421 | 0.0 | True | -323.015766 | -0.071217 | -0.008246 | 73.507898 | 0.0 | True | -0.102162 | -0.019793 | -0.054186 | 96.454575 | 0.0 | True | -3.015934 | -0.056306 | -0.086558 | 96.459236 | 0.0 | True | -0.000194 | -0.053865 | -0.062622 | 78.184954 | 0.0 | True |
| lag9 | -0.005669 | -0.107185 | -0.007692 | 100.733605 | 0.0 | True | -754.021483 | -0.112369 | -0.042993 | 77.162668 | 0.0 | True | 617.704521 | -0.051586 | 0.034847 | 85.785081 | 0.0 | True | -0.373157 | -0.101926 | -0.019951 | 78.419276 | 0.0 | True | -452.636236 | -0.082865 | -0.024129 | 68.105222 | 0.0 | True | -107.440313 | -0.122749 | -0.042393 | 104.111781 | 0.0 | True | -262.008726 | -0.058037 | -0.016566 | 98.876042 | 0.0 | True | -1.134204 | -0.045852 | -0.003525 | 76.305154 | 0.0 | True | -1058.862624 | -0.107398 | -0.035319 | 75.600919 | 0.0 | True | 315.975468 | -0.037052 | 0.008066 | 74.615339 | 0.0 | True | -0.056895 | -0.02511 | -0.030177 | 100.596646 | 0.0 | True | -1.970703 | -0.036607 | -0.05656 | 98.455342 | 0.0 | True | -0.00008 | -0.013841 | -0.025931 | 78.721289 | 0.0 | True |
| lag10 | 0.083026 | 0.017118 | 0.112653 | 102.231016 | 0.0 | True | 1004.003358 | -0.014985 | 0.057246 | 77.412537 | 0.0 | True | 469.670873 | -0.046375 | 0.026496 | 85.96606 | 0.0 | True | 1.199326 | -0.024556 | 0.064123 | 82.924341 | 0.0 | True | 778.922189 | -0.023311 | 0.041522 | 69.13319 | 0.0 | True | 279.258123 | 0.015387 | 0.110188 | 104.115985 | 0.0 | True | -410.990413 | -0.051631 | -0.025985 | 101.055261 | 0.0 | True | -3.432637 | -0.052928 | -0.010667 | 76.390433 | 0.0 | True | 1470.24773 | -0.031025 | 0.049041 | 76.965445 | 0.0 | True | 1181.147719 | -0.010737 | 0.030152 | 75.050302 | 0.0 | True | -0.109943 | -0.06343 | -0.058313 | 109.140713 | 0.0 | True | -1.410471 | -0.044812 | -0.040481 | 107.262913 | 0.0 | True | -0.000065 | -0.034233 | -0.020983 | 84.243052 | 0.0 | True |
| lag11 | -0.02583 | -0.100605 | -0.035047 | 102.23351 | 0.0 | True | 251.084721 | -0.04574 | 0.014316 | 77.425915 | 0.0 | True | 215.974668 | -0.031639 | 0.012184 | 87.375121 | 0.0 | True | 1.136972 | -0.000907 | 0.060789 | 82.947067 | 0.0 | True | 544.7335 | -0.024077 | 0.029038 | 69.145678 | 0.0 | True | 4.706126 | -0.064609 | 0.001857 | 104.126419 | 0.0 | True | -668.706701 | -0.070023 | -0.042279 | 101.150002 | 0.0 | True | -2.691346 | -0.051539 | -0.008364 | 77.14217 | 0.0 | True | 1003.004937 | -0.026702 | 0.033456 | 77.28177 | 0.0 | True | 739.926393 | -0.008984 | 0.018889 | 76.941524 | 0.0 | True | -0.157837 | -0.084849 | -0.083716 | 110.243408 | 0.0 | True | -2.961547 | -0.089641 | -0.084997 | 108.024662 | 0.0 | True | -0.000208 | -0.07406 | -0.0673 | 84.336634 | 0.0 | True |
| lag12 | 0.001054 | -0.041761 | 0.00143 | 106.35651 | 0.0 | True | -58.073707 | -0.045762 | -0.003311 | 77.459864 | 0.0 | True | -602.382299 | -0.069774 | -0.033983 | 87.412476 | 0.0 | True | -0.080719 | -0.032004 | -0.004316 | 85.394601 | 0.0 | True | 60.012594 | -0.035708 | 0.003199 | 69.58815 | 0.0 | True | -7.411247 | -0.044892 | -0.002924 | 104.513109 | 0.0 | True | 139.370819 | -0.027008 | 0.008812 | 103.028519 | 0.0 | True | -7.987301 | -0.070316 | -0.024821 | 77.853386 | 0.0 | True | 482.722219 | -0.022899 | 0.016101 | 77.934344 | 0.0 | True | -1542.238502 | -0.059435 | -0.03937 | 76.941759 | 0.0 | True | -0.056679 | -0.032902 | -0.030062 | 110.700119 | 0.0 | True | -0.870592 | -0.018887 | -0.024986 | 108.0327 | 0.0 | True | -0.000027 | 0.006718 | -0.008758 | 89.789509 | 0.0 | True |
| lag13 | 0.042825 | 0.025319 | 0.058106 | 110.953309 | 0.0 | True | 92.472266 | -0.033517 | 0.005273 | 78.483948 | 0.0 | True | 98.03982 | -0.020957 | 0.005531 | 88.126445 | 0.0 | True | -0.837336 | -0.054063 | -0.044769 | 85.830825 | 0.0 | True | -357.086393 | -0.043181 | -0.019035 | 70.044997 | 0.0 | True | 45.098732 | -0.018576 | 0.017795 | 110.198849 | 0.0 | True | 620.338871 | -0.006679 | 0.039221 | 103.385381 | 0.0 | True | 7.765795 | -0.021979 | 0.024133 | 78.005593 | 0.0 | True | -693.048156 | -0.045145 | -0.023117 | 78.083011 | 0.0 | True | 17.18481 | -0.003836 | 0.000439 | 78.451158 | 0.0 | True | -0.036462 | -0.046356 | -0.019339 | 111.009519 | 0.0 | True | 0.089392 | -0.019431 | 0.002566 | 108.443923 | 0.0 | True | 0.000207 | 0.044956 | 0.066823 | 89.811639 | 0.0 | True |
| lag14 | -0.045199 | -0.063115 | -0.061328 | 111.393957 | 0.0 | True | 507.675279 | 0.013786 | 0.028947 | 78.518265 | 0.0 | True | 428.432905 | -0.003022 | 0.02417 | 90.597783 | 0.0 | True | 0.353352 | 0.010033 | 0.018892 | 86.19516 | 0.0 | True | 362.688727 | 0.00552 | 0.019334 | 70.18412 | 0.0 | True | -172.859947 | -0.080022 | -0.068206 | 110.552445 | 0.0 | True | 270.264736 | -0.02299 | 0.017088 | 103.390537 | 0.0 | True | 3.591039 | -0.029964 | 0.01116 | 78.005598 | 0.0 | True | 330.653344 | 0.000755 | 0.011029 | 78.083053 | 0.0 | True | 1376.633632 | 0.022654 | 0.035143 | 78.625458 | 0.0 | True | -0.029998 | -0.038312 | -0.015911 | 112.155555 | 0.0 | True | -0.639122 | -0.05742 | -0.018343 | 109.349183 | 0.0 | True | -0.000013 | -0.043826 | -0.004255 | 90.150502 | 0.0 | True |
| lag15 | -0.013988 | -0.040208 | -0.01898 | 111.428295 | 0.0 | True | -92.895502 | -0.012295 | -0.005297 | 79.669205 | 0.0 | True | -796.75938 | -0.064129 | -0.044948 | 91.988842 | 0.0 | True | -0.322791 | -0.035363 | -0.017258 | 86.728023 | 0.0 | True | -200.062117 | -0.026241 | -0.010665 | 70.576211 | 0.0 | True | 43.089607 | -0.013612 | 0.017002 | 110.553911 | 0.0 | True | -32.47353 | -0.027092 | -0.002053 | 103.612112 | 0.0 | True | 0.02175 | -0.026421 | 0.000068 | 78.057782 | 0.0 | True | -5.576379 | -0.007672 | -0.000186 | 78.612362 | 0.0 | True | -467.609822 | -0.023657 | -0.011937 | 80.088523 | 0.0 | True | -0.05771 | -0.050774 | -0.030609 | 112.604577 | 0.0 | True | 0.947871 | 0.015384 | 0.027204 | 115.9993 | 0.0 | True | 0.000052 | 0.032389 | 0.016644 | 92.065404 | 0.0 | True |
| lag16 | -0.003903 | -0.042908 | -0.005296 | 111.441938 | 0.0 | True | -537.749153 | -0.034263 | -0.030661 | 80.750401 | 0.0 | True | -597.518554 | -0.067769 | -0.033708 | 92.386023 | 0.0 | True | -0.390208 | -0.042956 | -0.020863 | 86.988543 | 0.0 | True | -335.719634 | -0.033334 | -0.017896 | 71.261153 | 0.0 | True | -2.773208 | -0.041998 | -0.001094 | 110.563281 | 0.0 | True | -212.782355 | -0.023968 | -0.013453 | 104.684496 | 0.0 | True | -2.100902 | -0.033475 | -0.006529 | 78.287831 | 0.0 | True | -623.385182 | -0.029908 | -0.020793 | 79.387489 | 0.0 | True | -1354.202201 | -0.039232 | -0.03457 | 81.076499 | 0.0 | True | 0.036108 | -0.00276 | 0.019151 | 116.04555 | 0.0 | True | 2.567998 | 0.035576 | 0.073702 | 120.261141 | 0.0 | True | 0.000123 | 0.019931 | 0.039549 | 92.474573 | 0.0 | True |
| lag17 | 0.002459 | -0.024515 | 0.003337 | 111.507901 | 0.0 | True | -520.982662 | -0.046382 | -0.029705 | 81.628516 | 0.0 | True | 319.146792 | -0.029053 | 0.018004 | 93.88877 | 0.0 | True | -0.272726 | -0.043529 | -0.014582 | 86.990441 | 0.0 | True | -443.535355 | -0.046899 | -0.023643 | 71.29363 | 0.0 | True | -7.008379 | -0.038599 | -0.002765 | 110.577085 | 0.0 | True | -467.915179 | -0.035318 | -0.029584 | 105.559384 | 0.0 | True | 4.40927 | -0.000952 | 0.013702 | 78.341814 | 0.0 | True | -754.059503 | -0.048256 | -0.025152 | 79.422532 | 0.0 | True | -1112.352029 | -0.032747 | -0.028396 | 81.969453 | 0.0 | True | 0.099914 | 0.010599 | 0.052994 | 131.688717 | 0.0 | True | 2.054926 | 0.00965 | 0.058977 | 126.397313 | 0.0 | True | 0.000057 | 0.009135 | 0.018274 | 95.007137 | 0.0 | True |
| lag18 | 0.005405 | -0.023754 | 0.007334 | 112.116342 | 0.0 | True | -469.314542 | -0.049608 | -0.026759 | 83.632758 | 0.0 | True | 620.520882 | -0.007351 | 0.035006 | 94.239334 | 0.0 | True | -0.023264 | -0.026835 | -0.001244 | 87.00744 | 0.0 | True | -96.538758 | -0.025899 | -0.005146 | 71.378366 | 0.0 | True | -8.503021 | -0.040634 | -0.003355 | 110.58438 | 0.0 | True | -422.460281 | -0.040433 | -0.02671 | 106.510153 | 0.0 | True | -2.135007 | -0.021639 | -0.006635 | 79.822462 | 0.0 | True | -160.26352 | -0.029593 | -0.005346 | 80.166657 | 0.0 | True | -1057.063361 | -0.044605 | -0.026985 | 83.609308 | 0.0 | True | 0.212943 | 0.071233 | 0.112944 | 132.653433 | 0.0 | True | 2.464697 | 0.039836 | 0.070738 | 127.072291 | 0.0 | True | -0.000141 | -0.051222 | -0.045444 | 95.025366 | 0.0 | True |
| lag19 | 0.01641 | -0.003794 | 0.022265 | 116.218409 | 0.0 | True | 708.729477 | 0.012293 | 0.04041 | 89.217032 | 0.0 | True | 299.580762 | -0.013921 | 0.016901 | 97.726018 | 0.0 | True | 0.069608 | -0.029493 | 0.003722 | 90.609001 | 0.0 | True | 155.872917 | -0.020799 | 0.008309 | 77.080708 | 0.0 | True | 6.178892 | -0.015629 | 0.002438 | 124.283561 | 0.0 | True | 440.214274 | 0.004286 | 0.027833 | 115.020234 | 0.0 | True | -11.17677 | -0.051953 | -0.034733 | 85.246967 | 0.0 | True | 738.204302 | 0.000503 | 0.024623 | 82.576744 | 0.0 | True | 1431.878943 | 0.013477 | 0.036553 | 93.309273 | 0.0 | True | 0.052859 | -0.028306 | 0.028036 | 133.850378 | 0.0 | True | 0.817103 | -0.014272 | 0.023451 | 127.478556 | 0.0 | True | 0.000012 | 0.023998 | 0.003854 | 98.340544 | 0.0 | True |
| lag20 | 0.04259 | 0.047286 | 0.057788 | 116.218553 | 0.0 | True | 1182.512564 | 0.042645 | 0.067425 | 89.218636 | 0.0 | True | 944.395373 | 0.048607 | 0.053277 | 98.586347 | 0.0 | True | 1.012752 | 0.02908 | 0.054148 | 91.512036 | 0.0 | True | 1278.142007 | 0.047613 | 0.068134 | 77.551126 | 0.0 | True | 267.640834 | 0.08186 | 0.105605 | 125.094278 | 0.0 | True | 1316.468336 | 0.059816 | 0.083234 | 119.263196 | 0.0 | True | 21.383924 | 0.047414 | 0.066453 | 86.097752 | 0.0 | True | 1327.966085 | 0.03089 | 0.044295 | 82.965659 | 0.0 | True | 3481.013047 | 0.062649 | 0.088863 | 95.284739 | 0.0 | True | 0.058854 | 0.010118 | 0.031216 | 134.770286 | 0.0 | True | 0.633656 | 0.009783 | 0.018186 | 127.481887 | 0.0 | True | 0.000161 | 0.044014 | 0.05195 | 103.766011 | 0.0 | True |
| lag21 | -0.000252 | 0.038909 | -0.000342 | 119.386155 | 0.0 | True | -20.028635 | 0.000085 | -0.001142 | 92.462395 | 0.0 | True | 468.917795 | 0.060819 | 0.026454 | 101.695437 | 0.0 | True | 0.506905 | 0.01523 | 0.027102 | 94.778064 | 0.0 | True | 366.953572 | 0.012295 | 0.019561 | 79.004828 | 0.0 | True | 65.081382 | 0.051234 | 0.02568 | 126.894258 | 0.0 | True | 929.169499 | 0.048538 | 0.058747 | 125.935322 | 0.0 | True | 8.46514 | 0.03119 | 0.026306 | 88.146075 | 0.0 | True | 533.230467 | 0.022956 | 0.017786 | 85.092802 | 0.0 | True | 1570.261988 | 0.029448 | 0.040085 | 103.298852 | 0.0 | True | 0.051573 | 0.003604 | 0.027354 | 134.778076 | 0.0 | True | 0.057351 | -0.001103 | 0.001646 | 130.487331 | 0.0 | True | 0.000206 | 0.055946 | 0.066431 | 106.996038 | 0.0 | True |
| lag22 | 0.037395 | 0.104729 | 0.050738 | 124.553887 | 0.0 | True | 900.492749 | 0.082501 | 0.051344 | 97.514906 | 0.0 | True | 891.041049 | 0.130097 | 0.050267 | 101.889645 | 0.0 | True | 0.963609 | 0.075691 | 0.05152 | 99.99847 | 0.0 | True | 644.798049 | 0.059331 | 0.034372 | 83.602135 | 0.0 | True | 96.933166 | 0.10515 | 0.038247 | 128.99422 | 0.0 | True | 1164.686974 | 0.10088 | 0.073638 | 128.423228 | 0.0 | True | 13.129255 | 0.075061 | 0.040801 | 106.174367 | 0.0 | True | 1246.529192 | 0.073991 | 0.041578 | 93.276538 | 0.0 | True | 3161.417198 | 0.114938 | 0.080704 | 103.784814 | 0.0 | True | -0.004744 | -0.023927 | -0.002516 | 135.148608 | 0.0 | True | -1.722014 | -0.054936 | -0.049422 | 136.337897 | 0.0 | True | 0.000159 | 0.035862 | 0.051236 | 138.11918 | 0.0 | True |
| lag23 | 0.047743 | 0.148962 | 0.064779 | 412.743913 | 0.0 | True | 1123.378315 | 0.129563 | 0.064053 | 325.965764 | 0.0 | True | 222.603045 | 0.121787 | 0.012558 | 387.901269 | 0.0 | True | 1.217754 | 0.129267 | 0.065108 | 325.015217 | 0.0 | True | 1146.183748 | 0.109522 | 0.061099 | 298.497531 | 0.0 | True | 104.655193 | 0.135696 | 0.041294 | 417.912292 | 0.0 | True | 710.903382 | 0.108864 | 0.044947 | 356.290965 | 0.0 | True | 38.934505 | 0.189125 | 0.120994 | 372.159123 | 0.0 | True | 2443.973308 | 0.157568 | 0.081519 | 325.725552 | 0.0 | True | 778.165134 | 0.072124 | 0.019865 | 352.871297 | 0.0 | True | -0.032704 | -0.002426 | -0.017346 | 393.473719 | 0.0 | True | -2.401584 | -0.03469 | -0.068926 | 397.29468 | 0.0 | True | -0.000492 | -0.174069 | -0.158974 | 406.154378 | 0.0 | True |
| lag24 | -0.356382 | -0.452621 | -0.48355 | 420.460757 | 0.0 | True | -7550.66477 | -0.419618 | -0.430525 | 329.877006 | 0.0 | True | -8538.975666 | -0.454595 | -0.481719 | 401.883191 | 0.0 | True | -7.991557 | -0.414726 | -0.427277 | 330.410596 | 0.0 | True | -7833.077578 | -0.4122 | -0.417556 | 303.796284 | 0.0 | True | -1227.040315 | -0.443222 | -0.48416 | 418.752345 | 0.0 | True | -6800.665125 | -0.402005 | -0.429975 | 356.318099 | 0.0 | True | -149.486549 | -0.423435 | -0.464547 | 372.159192 | 0.0 | True | -13019.696769 | -0.409561 | -0.434276 | 332.245798 | 0.0 | True | -17610.108707 | -0.437084 | -0.449549 | 353.477729 | 0.0 | True | -0.863148 | -0.488434 | -0.45781 | 400.190162 | 0.0 | True | -16.032439 | -0.477307 | -0.460136 | 404.744811 | 0.0 | True | -0.001445 | -0.419304 | -0.466334 | 410.181786 | 0.0 | True |
| lag25 | 0.058292 | 0.024763 | 0.079093 | 431.719297 | 0.0 | True | 987.557305 | 0.017742 | 0.056309 | 341.095589 | 0.0 | True | 1887.181121 | 0.050046 | 0.106464 | 416.61146 | 0.0 | True | 1.236947 | 0.019143 | 0.066135 | 340.631363 | 0.0 | True | 1229.481226 | 0.045266 | 0.06554 | 312.615486 | 0.0 | True | 66.136452 | -0.006854 | 0.026096 | 435.752164 | 0.0 | True | 74.179503 | 0.103663 | 0.00469 | 358.694602 | 0.0 | True | -0.076192 | -0.059727 | -0.000237 | 391.111547 | 0.0 | True | 2179.642199 | -0.002851 | 0.072703 | 340.436465 | 0.0 | True | -868.548226 | 0.037217 | -0.022172 | 358.357519 | 0.0 | True | -0.13912 | 0.121221 | -0.073788 | 400.322818 | 0.0 | True | -2.707781 | 0.114219 | -0.077714 | 404.764392 | 0.0 | True | -0.000177 | 0.104213 | -0.057139 | 412.67764 | 0.0 | True |
| lag26 | 0.07038 | -0.099986 | 0.095494 | 436.762719 | 0.0 | True | 1671.822098 | -0.035145 | 0.095324 | 344.066964 | 0.0 | True | 1936.074401 | -0.084515 | 0.109222 | 420.329892 | 0.0 | True | 1.701759 | -0.036094 | 0.090986 | 344.702081 | 0.0 | True | 1585.498361 | -0.024489 | 0.084518 | 315.887553 | 0.0 | True | 297.389683 | -0.063226 | 0.117343 | 438.931762 | 0.0 | True | 693.923115 | -0.024795 | 0.043874 | 360.480973 | 0.0 | True | 39.869206 | -0.037988 | 0.123898 | 393.224036 | 0.0 | True | 2441.905853 | -0.043888 | 0.08145 | 341.068273 | 0.0 | True | 2462.74697 | -0.029826 | 0.062869 | 364.529127 | 0.0 | True | 0.019543 | -0.022195 | 0.010366 | 403.098863 | 0.0 | True | -0.13876 | -0.043683 | -0.003982 | 411.226531 | 0.0 | True | 0.000139 | -0.087692 | 0.044962 | 412.689431 | 0.0 | True |
| lag27 | 0.047085 | -0.046587 | 0.063887 | 449.789847 | 0.0 | True | 860.034103 | -0.029681 | 0.049038 | 349.986166 | 0.0 | True | 972.393428 | -0.047576 | 0.054857 | 430.399444 | 0.0 | True | 1.073514 | -0.034554 | 0.057396 | 347.693129 | 0.0 | True | 965.335076 | -0.026683 | 0.051459 | 320.001678 | 0.0 | True | 128.559963 | -0.061613 | 0.050727 | 450.276081 | 0.0 | True | 601.373049 | -0.052974 | 0.038022 | 365.276269 | 0.0 | True | 13.305132 | -0.036918 | 0.041347 | 404.095385 | 0.0 | True | 677.919149 | -0.070409 | 0.022612 | 348.864195 | 0.0 | True | 2768.434889 | -0.046495 | 0.070672 | 369.085458 | 0.0 | True | 0.089364 | 0.024872 | 0.047398 | 403.624252 | 0.0 | True | 2.519716 | 0.030062 | 0.072317 | 411.630787 | 0.0 | True | -0.00001 | -0.0335 | -0.003089 | 415.259549 | 0.0 | True |
| lag28 | 0.075642 | -0.015124 | 0.102634 | 450.041343 | 0.0 | True | 1213.344989 | -0.029707 | 0.069183 | 350.503452 | 0.0 | True | 1599.49513 | -0.028337 | 0.090234 | 430.432369 | 0.0 | True | 0.919813 | -0.026256 | 0.049179 | 348.939204 | 0.0 | True | 1081.985769 | -0.028869 | 0.057677 | 320.775292 | 0.0 | True | 242.730644 | -0.058426 | 0.095776 | 450.38083 | 0.0 | True | 984.876135 | -0.054993 | 0.062269 | 365.309291 | 0.0 | True | 30.170281 | -0.028259 | 0.093758 | 404.113451 | 0.0 | True | 2380.316329 | -0.019499 | 0.079396 | 349.466105 | 0.0 | True | 2377.707662 | -0.044975 | 0.060698 | 369.447378 | 0.0 | True | -0.03886 | -0.039458 | -0.020611 | 410.285534 | 0.0 | True | -0.629952 | -0.036485 | -0.01808 | 419.823523 | 0.0 | True | 0.000141 | 0.030612 | 0.045587 | 418.832934 | 0.0 | True |
| lag29 | -0.010506 | -0.028202 | -0.014254 | 450.833986 | 0.0 | True | -358.536828 | -0.04921 | -0.020443 | 350.543475 | 0.0 | True | 91.423217 | 0.001276 | 0.005158 | 435.029228 | 0.0 | True | -0.593438 | -0.022523 | -0.031729 | 348.970355 | 0.0 | True | -468.986566 | -0.025237 | -0.025 | 320.818638 | 0.0 | True | 23.314478 | -0.013656 | 0.009199 | 450.406625 | 0.0 | True | 81.693234 | -0.060633 | 0.005165 | 366.775548 | 0.0 | True | -1.229369 | 0.000973 | -0.00382 | 405.671822 | 0.0 | True | -661.122568 | -0.042437 | -0.022052 | 349.476297 | 0.0 | True | 669.843058 | -0.01474 | 0.0171 | 370.226508 | 0.0 | True | 0.138312 | 0.035896 | 0.07336 | 416.519781 | 0.0 | True | 2.834714 | 0.023005 | 0.081357 | 421.010356 | 0.0 | True | 0.000166 | 0.005018 | 0.053731 | 419.230951 | 0.0 | True |
| lag30 | -0.018643 | -0.026344 | -0.025295 | NaN | NaN | False | 99.686123 | -0.00217 | 0.005684 | NaN | NaN | False | -1079.790411 | -0.068419 | -0.060915 | NaN | NaN | False | 0.093789 | -0.019627 | 0.005015 | NaN | NaN | False | -110.965932 | -0.02355 | -0.005915 | NaN | NaN | False | -11.564539 | -0.062157 | -0.004563 | NaN | NaN | False | -544.139121 | -0.043441 | -0.034403 | NaN | NaN | False | -11.413114 | -0.025079 | -0.035468 | NaN | NaN | False | -85.992525 | -0.016255 | -0.002868 | NaN | NaN | False | 982.396528 | -0.024693 | 0.025078 | NaN | NaN | False | 0.133749 | -0.031523 | 0.07094 | NaN | NaN | False | 1.078462 | -0.035088 | 0.030952 | NaN | NaN | False | 0.000056 | 0.000361 | 0.017925 | NaN | NaN | False |












Auto-correlation/covariance structure



















































Stationary Analysis

| adf | kpss | |||||||
|---|---|---|---|---|---|---|---|---|
| stats | pval | number_of_used_lags | adf stationary | stats | pval | number_of_used_lags | kpss stationary | |
| CO(GT) | -24.900195 | 0.0 | 27 | True | 0.067891 | 0.1 | 123 | True |
| PT08.S1(CO) | -25.204788 | 0.0 | 32 | True | 0.064395 | 0.1 | 112 | True |
| NMHC(GT) | -10.008933 | 0.0 | 23 | True | 0.045934 | 0.1 | 45 | True |
| C6H6(GT) | -23.216466 | 0.0 | 30 | True | 0.027501 | 0.1 | 85 | True |
| PT08.S2(NMHC) | -23.845618 | 0.0 | 33 | True | 0.032222 | 0.1 | 105 | True |
| NOx(GT) | -25.052843 | 0.0 | 28 | True | 0.449359 | 0.05588 | 116 | True |
| PT08.S3(NOx) | -25.451232 | 0.0 | 34 | True | 0.051891 | 0.1 | 100 | True |
| NO2(GT) | -25.619444 | 0.0 | 29 | True | 0.199061 | 0.1 | 109 | True |
| PT08.S4(NO2) | -21.477915 | 0.0 | 34 | True | 0.033935 | 0.1 | 88 | True |
| PT08.S5(O3) | -24.778391 | 0.0 | 33 | True | 0.011084 | 0.1 | 94 | True |
| T | -22.534557 | 0.0 | 37 | True | 0.044319 | 0.1 | 54 | True |
| RH | -26.127431 | 0.0 | 31 | True | 0.007199 | 0.1 | 45 | True |
| AH | -29.005741 | 0.0 | 25 | True | 0.003645 | 0.1 | 17 | True |

| adf | kpss | |||||||
|---|---|---|---|---|---|---|---|---|
| stats | pval | number_of_used_lags | adf stationary | stats | pval | number_of_used_lags | kpss stationary | |
| CO(GT) | -26.931409 | 0.0 | 33 | True | 0.032225 | 0.1 | 196 | True |
| PT08.S1(CO) | -25.612593 | 0.0 | 34 | True | 0.013633 | 0.1 | 127 | True |
| NMHC(GT) | -10.750317 | 0.0 | 23 | True | 0.148104 | 0.1 | 257 | True |
| C6H6(GT) | -27.828471 | 0.0 | 32 | True | 0.027901 | 0.1 | 200 | True |
| PT08.S2(NMHC) | -26.544557 | 0.0 | 33 | True | 0.023625 | 0.1 | 150 | True |
| NOx(GT) | -25.496917 | 0.0 | 35 | True | 0.010477 | 0.1 | 123 | True |
| PT08.S3(NOx) | -26.476556 | 0.0 | 33 | True | 0.010565 | 0.1 | 89 | True |
| NO2(GT) | -24.272786 | 0.0 | 36 | True | 0.025971 | 0.1 | 126 | True |
| PT08.S4(NO2) | -26.402711 | 0.0 | 33 | True | 0.02212 | 0.1 | 145 | True |
| PT08.S5(O3) | -25.699396 | 0.0 | 33 | True | 0.008484 | 0.1 | 108 | True |
| T | -23.662499 | 0.0 | 37 | True | 0.005056 | 0.1 | 62 | True |
| RH | -26.636483 | 0.0 | 33 | True | 0.003763 | 0.1 | 49 | True |
| AH | -24.410589 | 0.0 | 34 | True | 0.001213 | 0.1 | 25 | True |

| adf | kpss | |||||||
|---|---|---|---|---|---|---|---|---|
| stats | pval | number_of_used_lags | adf stationary | stats | pval | number_of_used_lags | kpss stationary | |
| CO(GT) | -10.267951 | 0.0 | 37 | True | 1.326229 | 0.01 | 42 | False |
| PT08.S1(CO) | -9.701349 | 0.0 | 38 | True | 0.491353 | 0.043614 | 47 | False |
| NMHC(GT) | -2.937991 | 0.041105 | 23 | True | 1.176806 | 0.01 | 16 | False |
| C6H6(GT) | -10.300587 | 0.0 | 37 | True | 0.830241 | 0.01 | 42 | False |
| PT08.S2(NMHC) | -10.395797 | 0.0 | 37 | True | 1.042781 | 0.01 | 43 | False |
| NOx(GT) | -6.883601 | 0.0 | 26 | True | 9.428993 | 0.01 | 50 | False |
| PT08.S3(NOx) | -10.581876 | 0.0 | 36 | True | 3.567417 | 0.01 | 49 | False |
| NO2(GT) | -7.526296 | 0.0 | 32 | True | 8.06908 | 0.01 | 48 | False |
| PT08.S4(NO2) | -5.966929 | 0.0 | 37 | True | 9.684327 | 0.01 | 53 | False |
| PT08.S5(O3) | -11.032393 | 0.0 | 36 | True | 1.065825 | 0.01 | 48 | False |
| T | -2.883684 | 0.047278 | 29 | True | 7.578053 | 0.01 | 55 | False |
| RH | -7.3299 | 0.0 | 38 | True | 3.12812 | 0.01 | 52 | False |
| AH | -5.069178 | 0.000016 | 25 | True | 4.324071 | 0.01 | 57 | False |
Exploratory Data Analysis
Exploratory for time series process
Deterministic process exploration of time series features

| CO(GT) | PT08.S1(CO) | NMHC(GT) | C6H6(GT) | PT08.S2(NMHC) | NOx(GT) | PT08.S3(NOx) | NO2(GT) | PT08.S4(NO2) | PT08.S5(O3) | T | RH | AH | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| coef | P>|z| | reject | coef | P>|z| | reject | coef | P>|z| | reject | coef | P>|z| | reject | coef | P>|z| | reject | coef | P>|z| | reject | coef | P>|z| | reject | coef | P>|z| | reject | coef | P>|z| | reject | coef | P>|z| | reject | coef | P>|z| | reject | coef | P>|z| | reject | coef | P>|z| | reject | |
| const | 1.8668 | 0.000 | True | 1122.5781 | 0.000 | True | 132.3782 | 0.000 | True | 8.8940 | 0.000 | True | 881.3584 | 0.000 | True | 47.5578 | 0.000 | True | 978.3144 | 0.000 | True | 94.8901 | 0.000 | True | 1567.2992 | 0.000 | True | 903.3162 | 0.000 | True | 16.5477 | 0.000 | True | 42.3671 | 0.000 | True | 0.7507 | 0.000 | True |
| trend | 5.793e-05 | 0.001 | True | -0.0393 | 0.000 | True | 0.1886 | 0.000 | True | 0.0010 | 0.000 | True | 0.0353 | 0.000 | True | 0.0555 | 0.000 | True | -0.0673 | 0.000 | True | -0.0063 | 0.000 | True | 0.0290 | 0.000 | True | 0.0256 | 0.000 | True | 0.0038 | 0.000 | True | 0.0006 | 0.004 | True | 0.0003 | 0.000 | True |
| trend_squared | -2.833e-09 | 0.107 | False | 4.175e-06 | 0.000 | True | -6.751e-05 | 0.048 | True | -1.24e-07 | 0.000 | True | -4.685e-06 | 0.000 | True | -2.406e-06 | 0.000 | True | 4.604e-06 | 0.000 | True | 1.361e-06 | 0.000 | True | -1.041e-05 | 0.000 | True | -1.055e-06 | 0.055 | False | -5.67e-07 | 0.000 | True | 8.179e-08 | 0.000 | True | -3.241e-08 | 0.000 | True |
| sin(1,24) | -0.3435 | 0.000 | True | -58.8431 | 0.000 | True | -76.0408 | 0.000 | True | -2.3799 | 0.000 | True | -93.3787 | 0.000 | True | -58.0096 | 0.000 | True | 66.7070 | 0.000 | True | -13.9948 | 0.000 | True | -71.3560 | 0.000 | True | -67.4841 | 0.000 | True | -2.6439 | 0.000 | True | 7.2555 | 0.000 | True | 0.0216 | 0.000 | True |
| cos(1,24) | 0.7431 | 0.000 | True | 101.4581 | 0.000 | True | 79.3568 | 0.000 | True | 3.4164 | 0.000 | True | 144.5080 | 0.000 | True | 61.1878 | 0.000 | True | -95.3206 | 0.000 | True | 26.6262 | 0.000 | True | 107.6307 | 0.000 | True | 114.2243 | 0.000 | True | 3.4038 | 0.000 | True | -8.4095 | 0.000 | True | -0.0074 | 0.201 | False |
| sin(2,24) | 0.5671 | 0.000 | True | 89.1333 | 0.000 | True | 78.7232 | 0.000 | True | 3.0023 | 0.000 | True | 116.7752 | 0.000 | True | 67.7697 | 0.000 | True | -102.1578 | 0.000 | True | 17.1826 | 0.000 | True | 111.0971 | 0.000 | True | 177.0372 | 0.000 | True | -0.8017 | 0.000 | True | 1.7592 | 0.000 | True | 0.0140 | 0.001 | True |
| cos(2,24) | 0.2356 | 0.000 | True | 33.7553 | 0.000 | True | 50.1046 | 0.000 | True | 1.4764 | 0.000 | True | 42.6933 | 0.000 | True | 12.9467 | 0.000 | True | 11.4231 | 0.002 | True | 1.1648 | 0.056 | False | 64.7984 | 0.000 | True | 29.0433 | 0.000 | True | -0.4683 | 0.000 | True | 2.3813 | 0.000 | True | 0.0042 | 0.465 | False |
| sin(3,24) | -0.0580 | 0.000 | True | -26.0255 | 0.000 | True | -7.6270 | 0.129 | False | -0.7653 | 0.000 | True | -34.5946 | 0.000 | True | -10.6502 | 0.000 | True | 36.6993 | 0.000 | True | -2.3773 | 0.000 | True | -31.3248 | 0.000 | True | -31.7965 | 0.000 | True | 0.1074 | 0.188 | False | -0.4219 | 0.026 | True | 0.0009 | 0.828 | False |
| cos(3,24) | 0.3342 | 0.000 | True | 49.4504 | 0.000 | True | 39.6495 | 0.000 | True | 1.6929 | 0.000 | True | 60.9457 | 0.000 | True | 32.1642 | 0.000 | True | -20.3776 | 0.000 | True | 8.9774 | 0.000 | True | 66.9855 | 0.000 | True | 77.1826 | 0.000 | True | 0.4575 | 0.000 | True | -0.3176 | 0.218 | False | 0.0127 | 0.029 | True |
| s(2,24) | 0.2564 | 0.000 | True | 40.9947 | 0.001 | True | 73.0667 | 0.003 | True | 1.5034 | 0.000 | True | 52.9256 | 0.000 | True | 13.9287 | 0.181 | False | -27.5242 | 0.041 | True | 3.9521 | 0.070 | False | 57.2217 | 0.000 | True | 56.0048 | 0.014 | True | -0.2041 | 0.608 | False | 0.8021 | 0.384 | False | 0.0072 | 0.728 | False |
| s(3,24) | 0.2972 | 0.000 | True | 51.2380 | 0.000 | True | 57.6958 | 0.003 | True | 1.0449 | 0.001 | True | 53.5165 | 0.000 | True | 24.3747 | 0.003 | True | -14.6088 | 0.175 | False | 6.2627 | 0.000 | True | 63.2296 | 0.000 | True | 68.5426 | 0.000 | True | 0.1012 | 0.750 | False | 1.4206 | 0.054 | False | 0.0197 | 0.236 | False |
| s(4,24) | -0.0395 | 0.413 | False | 18.8420 | 0.016 | True | -34.9657 | 0.030 | True | -0.7628 | 0.003 | True | 1.7356 | 0.845 | False | 6.8634 | 0.320 | False | 45.7242 | 0.000 | True | 3.9174 | 0.007 | True | 21.7835 | 0.043 | True | 11.4940 | 0.447 | False | 0.5425 | 0.039 | True | 1.6276 | 0.008 | True | 0.0279 | 0.042 | True |
| s(5,24) | -0.1629 | 0.001 | True | 22.6352 | 0.004 | True | -32.6858 | 0.042 | True | -0.6655 | 0.010 | True | 1.2761 | 0.885 | False | -18.9986 | 0.006 | True | 79.2625 | 0.000 | True | 0.5924 | 0.681 | False | 36.2809 | 0.001 | True | -0.0954 | 0.995 | False | 0.8774 | 0.001 | True | 1.6515 | 0.007 | True | 0.0342 | 0.012 | True |
| s(6,24) | 0.0659 | 0.183 | False | 56.5781 | 0.000 | True | -0.1038 | 0.995 | False | 0.3905 | 0.140 | False | 38.4212 | 0.000 | True | -9.4056 | 0.184 | False | 59.9567 | 0.000 | True | 3.3868 | 0.022 | True | 74.8513 | 0.000 | True | 39.2220 | 0.012 | True | 0.8823 | 0.001 | True | 2.0498 | 0.001 | True | 0.0383 | 0.007 | True |
| s(7,24) | 0.2010 | 0.000 | True | 68.1414 | 0.000 | True | 6.2004 | 0.700 | False | 0.8740 | 0.001 | True | 58.0034 | 0.000 | True | 7.0059 | 0.309 | False | 32.2051 | 0.000 | True | 7.0615 | 0.000 | True | 86.9150 | 0.000 | True | 68.4885 | 0.000 | True | 0.6548 | 0.013 | True | 2.3737 | 0.000 | True | 0.0378 | 0.006 | True |
| s(8,24) | 0.0961 | 0.043 | True | 44.9320 | 0.000 | True | 3.5582 | 0.823 | False | 0.3306 | 0.191 | False | 39.2707 | 0.000 | True | 0.9803 | 0.885 | False | 25.3600 | 0.004 | True | 3.8039 | 0.007 | True | 63.9781 | 0.000 | True | 39.9341 | 0.007 | True | 0.3695 | 0.153 | False | 2.5238 | 0.000 | True | 0.0332 | 0.014 | True |
| s(9,24) | -0.0388 | 0.421 | False | 28.6848 | 0.000 | True | 3.3932 | 0.834 | False | -0.0688 | 0.789 | False | 19.3427 | 0.029 | True | -7.6752 | 0.266 | False | 35.6000 | 0.000 | True | -0.2900 | 0.841 | False | 49.0774 | 0.000 | True | 16.0293 | 0.289 | False | 0.1818 | 0.490 | False | 2.6502 | 0.000 | True | 0.0301 | 0.028 | True |
| s(10,24) | -0.0353 | 0.465 | False | 29.1878 | 0.000 | True | 3.6199 | 0.824 | False | -0.0232 | 0.928 | False | 14.9397 | 0.091 | False | -4.2656 | 0.536 | False | 53.2641 | 0.000 | True | -0.0069 | 0.996 | False | 48.1151 | 0.000 | True | 14.9726 | 0.322 | False | 0.1829 | 0.487 | False | 2.5435 | 0.000 | True | 0.0271 | 0.048 | True |
| s(11,24) | 0.0245 | 0.606 | False | 28.3382 | 0.000 | True | -8.2052 | 0.607 | False | -0.1370 | 0.588 | False | 7.9735 | 0.359 | False | -8.1450 | 0.230 | False | 73.5606 | 0.000 | True | -0.6106 | 0.667 | False | 43.3273 | 0.000 | True | 12.5888 | 0.397 | False | 0.3983 | 0.124 | False | 2.3685 | 0.000 | True | 0.0301 | 0.025 | True |
| s(12,24) | -0.1061 | 0.026 | True | 14.5048 | 0.060 | False | -30.0302 | 0.061 | False | -0.8586 | 0.001 | True | -14.2112 | 0.104 | False | -16.7454 | 0.014 | True | 95.6318 | 0.000 | True | -0.9151 | 0.521 | False | 21.6749 | 0.041 | True | -8.5199 | 0.568 | False | 0.6743 | 0.009 | True | 2.1046 | 0.000 | True | 0.0347 | 0.010 | True |
| s(13,24) | -0.1988 | 0.000 | True | 10.1817 | 0.192 | False | -48.7850 | 0.003 | True | -1.4119 | 0.000 | True | -15.0520 | 0.088 | False | -24.2334 | 0.000 | True | 72.7188 | 0.000 | True | 0.1333 | 0.926 | False | 10.7292 | 0.317 | False | -14.4253 | 0.339 | False | 0.6124 | 0.020 | True | 2.6206 | 0.000 | True | 0.0351 | 0.010 | True |
| s(14,24) | 0.0958 | 0.044 | True | 75.1439 | 0.000 | True | 16.6751 | 0.298 | False | 1.0258 | 0.000 | True | 69.0490 | 0.000 | True | 0.8175 | 0.904 | False | 11.6361 | 0.185 | False | 4.0162 | 0.005 | True | 102.0455 | 0.000 | True | 63.6718 | 0.000 | True | 0.3957 | 0.128 | False | 2.9181 | 0.000 | True | 0.0331 | 0.015 | True |
| s(15,24) | 0.4543 | 0.000 | True | 110.1493 | 0.000 | True | 99.8860 | 0.000 | True | 3.4901 | 0.000 | True | 134.0978 | 0.000 | True | 41.4312 | 0.000 | True | -22.1986 | 0.011 | True | 9.0113 | 0.000 | True | 179.2762 | 0.000 | True | 126.9461 | 0.000 | True | 0.1641 | 0.526 | False | 2.9720 | 0.000 | True | 0.0307 | 0.023 | True |
| s(16,24) | 0.2554 | 0.000 | True | 44.7241 | 0.000 | True | 33.7237 | 0.038 | True | 0.7617 | 0.003 | True | 41.7777 | 0.000 | True | 17.6604 | 0.010 | True | 33.2547 | 0.000 | True | 4.4187 | 0.002 | True | 64.5298 | 0.000 | True | 58.8152 | 0.000 | True | 0.1058 | 0.688 | False | 2.5842 | 0.000 | True | 0.0315 | 0.022 | True |
| s(17,24) | -0.1537 | 0.001 | True | 1.6535 | 0.832 | False | -46.0848 | 0.005 | True | -1.5666 | 0.000 | True | -30.0305 | 0.001 | True | -9.2514 | 0.180 | False | 77.0868 | 0.000 | True | 1.6529 | 0.252 | False | -10.8964 | 0.311 | False | -5.5178 | 0.715 | False | 0.8844 | 0.001 | True | 0.5469 | 0.370 | False | 0.0285 | 0.038 | True |
| s(18,24) | -0.2001 | 0.000 | True | 15.9993 | 0.037 | True | -47.4870 | 0.003 | True | -1.1453 | 0.000 | True | -11.2676 | 0.195 | False | -12.0484 | 0.075 | False | 66.7953 | 0.000 | True | 2.3077 | 0.103 | False | 10.1024 | 0.339 | False | -9.7901 | 0.510 | False | 1.2089 | 0.000 | True | 0.2972 | 0.620 | False | 0.0252 | 0.061 | False |
| s(19,24) | 0.0665 | 0.168 | False | 64.8712 | 0.000 | True | 8.6532 | 0.593 | False | 0.8015 | 0.002 | True | 58.5529 | 0.000 | True | 0.1879 | 0.978 | False | 28.5310 | 0.001 | True | 5.6053 | 0.000 | True | 88.1275 | 0.000 | True | 45.0089 | 0.003 | True | 1.2493 | 0.000 | True | 0.9491 | 0.119 | False | 0.0322 | 0.019 | True |
| s(20,24) | 0.3731 | 0.000 | True | 99.6662 | 0.000 | True | 62.0683 | 0.000 | True | 2.4092 | 0.000 | True | 112.3967 | 0.000 | True | 19.3744 | 0.006 | True | 0.5886 | 0.949 | False | 8.7930 | 0.000 | True | 147.1244 | 0.000 | True | 101.2843 | 0.000 | True | 1.0363 | 0.000 | True | 1.7462 | 0.005 | True | 0.0378 | 0.007 | True |
| s(21,24) | 0.3506 | 0.000 | True | 74.9109 | 0.000 | True | 41.4635 | 0.011 | True | 1.7050 | 0.000 | True | 82.3891 | 0.000 | True | 22.8307 | 0.001 | True | 21.6825 | 0.015 | True | 8.2331 | 0.000 | True | 113.6375 | 0.000 | True | 77.0806 | 0.000 | True | 0.7163 | 0.006 | True | 1.7301 | 0.005 | True | 0.0327 | 0.017 | True |
| s(22,24) | 0.0317 | 0.512 | False | 31.0755 | 0.000 | True | 9.2083 | 0.571 | False | -0.0420 | 0.871 | False | 19.4798 | 0.028 | True | 2.7642 | 0.689 | False | 45.2817 | 0.000 | True | 3.5543 | 0.014 | True | 40.9945 | 0.000 | True | 22.6438 | 0.135 | False | 0.8457 | 0.001 | True | 0.6551 | 0.284 | False | 0.0243 | 0.077 | False |
| s(23,24) | -0.2435 | 0.000 | True | -14.6270 | 0.122 | False | -38.4017 | 0.050 | False | -1.8100 | 0.000 | True | -44.3346 | 0.000 | True | -15.6345 | 0.061 | False | 57.3659 | 0.000 | True | -1.8344 | 0.294 | False | -37.8063 | 0.004 | True | -45.9646 | 0.012 | True | 0.9636 | 0.002 | True | -0.6302 | 0.394 | False | 0.0126 | 0.447 | False |
| s(24,24) | -0.2477 | 0.001 | True | -23.6292 | 0.045 | True | -39.4959 | 0.106 | False | -1.4562 | 0.000 | True | -41.7626 | 0.002 | True | -13.8200 | 0.185 | False | 35.5303 | 0.008 | True | -1.9856 | 0.363 | False | -43.7367 | 0.007 | True | -46.8671 | 0.040 | True | 0.6421 | 0.106 | False | -0.9811 | 0.288 | False | -0.0007 | 0.972 | False |
| CO(GT) | PT08.S1(CO) | NMHC(GT) | C6H6(GT) | PT08.S2(NMHC) | NOx(GT) | PT08.S3(NOx) | NO2(GT) | PT08.S4(NO2) | PT08.S5(O3) | T | RH | AH | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| const | 1.866800 | 1122.5781 | 132.3782 | 8.894 | 881.3584 | 47.5578 | 978.3144 | 94.8901 | 1567.2992 | 903.3162 | 16.5477 | 42.3671 | 0.7507 |
| trend | 0.000058 | -0.0393 | 0.1886 | 0.001 | 0.0353 | 0.0555 | -0.0673 | -0.0063 | 0.0290 | 0.0256 | 0.0038 | 0.0006 | 0.0003 |
| CO(GT) | PT08.S1(CO) | NMHC(GT) | C6H6(GT) | PT08.S2(NMHC) | NOx(GT) | PT08.S3(NOx) | NO2(GT) | PT08.S4(NO2) | PT08.S5(O3) | T | RH | AH | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| period | s(15,24) | s(15,24) | s(15,24) | s(15,24) | s(15,24) | s(15,24) | s(12,24) | s(15,24) | s(15,24) | s(15,24) | s(19,24) | s(15,24) | s(6,24) |
| coef | 0.4543 | 110.1493 | 99.886 | 3.4901 | 134.0978 | 41.4312 | 95.6318 | 9.0113 | 179.2762 | 126.9461 | 1.2493 | 2.972 | 0.0383 |
Stochastic process exploration of time series features

| CO(GT) | PT08.S1(CO) | NMHC(GT) | C6H6(GT) | PT08.S2(NMHC) | NOx(GT) | PT08.S3(NOx) | NO2(GT) | PT08.S4(NO2) | PT08.S5(O3) | T | RH | AH | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| coef | std err | z | P>|z| | [0.025 | 0.975] | coef | std err | z | P>|z| | [0.025 | 0.975] | coef | std err | z | P>|z| | [0.025 | 0.975] | coef | std err | z | P>|z| | [0.025 | 0.975] | coef | std err | z | P>|z| | [0.025 | 0.975] | coef | std err | z | P>|z| | [0.025 | 0.975] | coef | std err | z | P>|z| | [0.025 | 0.975] | coef | std err | z | P>|z| | [0.025 | 0.975] | coef | std err | z | P>|z| | [0.025 | 0.975] | coef | std err | z | P>|z| | [0.025 | 0.975] | coef | std err | z | P>|z| | [0.025 | 0.975] | coef | std err | z | P>|z| | [0.025 | 0.975] | coef | std err | z | P>|z| | [0.025 | 0.975] | |||
| model name | param symbol | param name | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| strend | $\\sigma_\\varepsilon^2$ | sigma2.irregular | 1.458e-09 | 0.007 | 2.23e-07 | 1.000 | -0.013 | 0.013 | 0.0002 | 96.132 | 2.12e-06 | 1.000 | -188.414 | 188.415 | 5.3e-14 | 418.307 | 1.27e-16 | 1.000 | -819.866 | 819.866 | 6.441e-08 | 0.169 | 3.8e-07 | 1.000 | -0.332 | 0.332 | 3.811e-05 | 170.628 | 2.23e-07 | 1.000 | -334.425 | 334.425 | 0.0238 | 55.537 | 0.000 | 1.000 | -108.826 | 108.874 | 0.0006 | 81.657 | 7.33e-06 | 1.000 | -160.044 | 160.045 | 19.8679 | 2.528 | 7.859 | 0.000 | 14.913 | 24.823 | 1.033e-10 | 216.925 | 4.76e-13 | 1.000 | -425.165 | 425.165 | 4.344e-06 | 260.182 | 1.67e-08 | 1.000 | -509.947 | 509.947 | 0.1006 | 0.010 | 10.400 | 0.000 | 0.082 | 0.120 | 0.1959 | 0.284 | 0.689 | 0.491 | -0.361 | 0.753 | 6.115e-10 | 1.79e-05 | 3.42e-05 | 1.000 | -3.5e-05 | 3.5e-05 |
| $\\sigma_\\eta^2$ | sigma2.trend | 4.735e-09 | 1.67e-09 | 2.836 | 0.005 | 1.46e-09 | 8.01e-09 | 0.8443 | 0.088 | 9.584 | 0.000 | 0.672 | 1.017 | 0.0472 | 0.018 | 2.634 | 0.008 | 0.012 | 0.082 | 0.0004 | 4.77e-05 | 8.511 | 0.000 | 0.000 | 0.000 | 0.7082 | 0.082 | 8.667 | 0.000 | 0.548 | 0.868 | 0.2031 | 0.018 | 11.182 | 0.000 | 0.168 | 0.239 | 2.0802 | 0.168 | 12.402 | 0.000 | 1.751 | 2.409 | 0.0135 | 0.001 | 10.088 | 0.000 | 0.011 | 0.016 | 1.4955 | 0.160 | 9.370 | 0.000 | 1.183 | 1.808 | 8.6737 | 0.740 | 11.726 | 0.000 | 7.224 | 10.123 | 0.6361 | 0.016 | 39.365 | 0.000 | 0.604 | 0.668 | 4.2851 | 0.158 | 27.163 | 0.000 | 3.976 | 4.594 | 3.49e-06 | 2.93e-07 | 11.909 | 0.000 | 2.92e-06 | 4.06e-06 | |
| $\\sigma_\\kappa^2$ | sigma2.cycle | 0.4720 | 0.012 | 39.577 | 0.000 | 0.449 | 0.495 | 7089.1688 | 185.434 | 38.230 | 0.000 | 6725.725 | 7452.613 | 1.019e+04 | 771.010 | 13.219 | 0.000 | 8680.668 | 1.17e+04 | 12.2176 | 0.315 | 38.824 | 0.000 | 11.601 | 12.834 | 1.238e+04 | 326.117 | 37.950 | 0.000 | 1.17e+04 | 1.3e+04 | 5502.7222 | 112.405 | 48.954 | 0.000 | 5282.412 | 5723.032 | 7745.2621 | 173.296 | 44.694 | 0.000 | 7405.608 | 8084.916 | 237.3133 | 5.214 | 45.517 | 0.000 | 227.094 | 247.532 | 1.403e+04 | 410.915 | 34.135 | 0.000 | 1.32e+04 | 1.48e+04 | 1.918e+04 | 527.359 | 36.366 | 0.000 | 1.81e+04 | 2.02e+04 | 0.1001 | 0.016 | 6.270 | 0.000 | 0.069 | 0.131 | 6.0336 | 0.658 | 9.169 | 0.000 | 4.744 | 7.323 | 0.0019 | 4.16e-05 | 46.691 | 0.000 | 0.002 | 0.002 | |
| $\\lambda_c$ | frequency.cycle | 0.2701 | 0.006 | 46.016 | 0.000 | 0.259 | 0.282 | 0.3524 | 0.005 | 64.570 | 0.000 | 0.342 | 0.363 | 0.3749 | 0.020 | 18.556 | 0.000 | 0.335 | 0.415 | 0.3503 | 0.006 | 57.485 | 0.000 | 0.338 | 0.362 | 0.3407 | 0.005 | 63.952 | 0.000 | 0.330 | 0.351 | 0.3067 | 0.004 | 72.813 | 0.000 | 0.298 | 0.315 | 0.3436 | 0.004 | 85.254 | 0.000 | 0.336 | 0.352 | 0.3148 | 0.004 | 81.049 | 0.000 | 0.307 | 0.322 | 0.3813 | 0.007 | 53.344 | 0.000 | 0.367 | 0.395 | 0.3891 | 0.005 | 73.227 | 0.000 | 0.379 | 0.399 | 1.0198 | 0.028 | 36.986 | 0.000 | 0.966 | 1.074 | 0.6238 | 0.041 | 15.094 | 0.000 | 0.543 | 0.705 | 0.1898 | 0.005 | 36.413 | 0.000 | 0.180 | 0.200 | |
| $\\rho$ | damping.cycle | 0.8483 | 0.005 | 160.053 | 0.000 | 0.838 | 0.859 | 0.8669 | 0.004 | 211.665 | 0.000 | 0.859 | 0.875 | 0.8110 | 0.016 | 51.880 | 0.000 | 0.780 | 0.842 | 0.8386 | 0.005 | 182.079 | 0.000 | 0.830 | 0.848 | 0.8719 | 0.004 | 218.422 | 0.000 | 0.864 | 0.880 | 0.8665 | 0.004 | 236.743 | 0.000 | 0.859 | 0.874 | 0.8873 | 0.004 | 239.450 | 0.000 | 0.880 | 0.895 | 0.8987 | 0.003 | 275.784 | 0.000 | 0.892 | 0.905 | 0.8324 | 0.005 | 172.457 | 0.000 | 0.823 | 0.842 | 0.8766 | 0.004 | 201.633 | 0.000 | 0.868 | 0.885 | 0.7724 | 0.022 | 35.327 | 0.000 | 0.730 | 0.815 | 0.7079 | 0.015 | 46.885 | 0.000 | 0.678 | 0.738 | 0.9099 | 0.003 | 286.604 | 0.000 | 0.904 | 0.916 | |
| $2 \\pi / \\lambda_c$ | period.cycle | 23.262441 | NaN | NaN | NaN | NaN | NaN | 17.829697 | NaN | NaN | NaN | NaN | NaN | 16.75963 | NaN | NaN | NaN | NaN | NaN | 17.936584 | NaN | NaN | NaN | NaN | NaN | 18.441988 | NaN | NaN | NaN | NaN | NaN | 20.486421 | NaN | NaN | NaN | NaN | NaN | 18.286337 | NaN | NaN | NaN | NaN | NaN | 19.959293 | NaN | NaN | NaN | NaN | NaN | 16.478325 | NaN | NaN | NaN | NaN | NaN | 16.147996 | NaN | NaN | NaN | NaN | NaN | 6.161194 | NaN | NaN | NaN | NaN | NaN | 10.072436 | NaN | NaN | NaN | NaN | NaN | 33.104243 | NaN | NaN | NaN | NaN | NaN | |
| lltrend | $\\sigma_\\varepsilon^2$ | sigma2.irregular | 3.739e-05 | 0.006 | 0.007 | 0.995 | -0.011 | 0.011 | 0.0195 | 103.117 | 0.000 | 1.000 | -202.086 | 202.126 | 2.117e-11 | 424.601 | 4.99e-14 | 1.000 | -832.204 | 832.204 | 1.591e-12 | 0.173 | 9.17e-12 | 1.000 | -0.340 | 0.340 | 2.965e-07 | 187.163 | 1.58e-09 | 1.000 | -366.833 | 366.833 | 0.2957 | 56.284 | 0.005 | 0.996 | -110.019 | 110.610 | 3.518e-05 | 81.704 | 4.31e-07 | 1.000 | -160.137 | 160.137 | 0.0098 | 3.276 | 0.003 | 0.998 | -6.412 | 6.431 | 9.3434 | 219.785 | 0.043 | 0.966 | -421.427 | 440.113 | 8.037e-11 | 225.024 | 3.57e-13 | 1.000 | -441.040 | 441.040 | 0.0875 | 0.027 | 3.236 | 0.001 | 0.035 | 0.141 | 0.0232 | 0.304 | 0.076 | 0.939 | -0.573 | 0.619 | 5.637e-12 | 1.51e-05 | 3.74e-07 | 1.000 | -2.95e-05 | 2.95e-05 |
| $\\sigma_\\zeta^2$ | sigma2.level | 0.0102 | 0.001 | 7.501 | 0.000 | 0.008 | 0.013 | 476.4186 | 51.422 | 9.265 | 0.000 | 375.633 | 577.205 | 143.4461 | 62.904 | 2.280 | 0.023 | 20.156 | 266.736 | 0.3678 | 0.049 | 7.530 | 0.000 | 0.272 | 0.464 | 530.3651 | 68.908 | 7.697 | 0.000 | 395.307 | 665.423 | 230.8066 | 22.391 | 10.308 | 0.000 | 186.921 | 274.692 | 856.0848 | 67.441 | 12.694 | 0.000 | 723.903 | 988.267 | 8.9299 | 1.155 | 7.734 | 0.000 | 6.667 | 11.193 | 937.4003 | 88.415 | 10.602 | 0.000 | 764.111 | 1110.690 | 9239.9681 | 562.973 | 16.413 | 0.000 | 8136.561 | 1.03e+04 | 0.0914 | 0.159 | 0.573 | 0.567 | -0.221 | 0.404 | 6.5201 | 2.599 | 2.509 | 0.012 | 1.426 | 11.614 | 0.0014 | 8.14e-05 | 17.393 | 0.000 | 0.001 | 0.002 | |
| $\\sigma_\\eta^2$ | sigma2.trend | 6.767e-13 | 1.02e-08 | 6.61e-05 | 1.000 | -2.01e-08 | 2.01e-08 | 2.055e-11 | 0.000 | 5.57e-08 | 1.000 | -0.001 | 0.001 | 1.047e-11 | 0.003 | 3.23e-09 | 1.000 | -0.006 | 0.006 | 1.282e-13 | 3.84e-07 | 3.34e-07 | 1.000 | -7.52e-07 | 7.52e-07 | 4.593e-10 | 0.001 | 8.83e-07 | 1.000 | -0.001 | 0.001 | 2.448e-07 | 0.000 | 0.002 | 0.998 | -0.000 | 0.000 | 1.792e-08 | 0.000 | 4.25e-05 | 1.000 | -0.001 | 0.001 | 1.637e-14 | 4e-06 | 4.09e-09 | 1.000 | -7.85e-06 | 7.85e-06 | 2.869e-05 | 0.001 | 0.030 | 0.976 | -0.002 | 0.002 | 5.058e-19 | 0.016 | 3.19e-17 | 1.000 | -0.031 | 0.031 | 0.6094 | 0.025 | 24.358 | 0.000 | 0.560 | 0.658 | 3.8793 | 0.229 | 16.926 | 0.000 | 3.430 | 4.329 | 4.399e-14 | 3.2e-09 | 1.38e-05 | 1.000 | -6.26e-09 | 6.26e-09 | |
| $\\sigma_\\kappa^2$ | sigma2.cycle | 0.3937 | 0.011 | 35.709 | 0.000 | 0.372 | 0.415 | 6696.9481 | 219.150 | 30.559 | 0.000 | 6267.421 | 7126.475 | 9708.6946 | 842.320 | 11.526 | 0.000 | 8057.778 | 1.14e+04 | 11.6647 | 0.353 | 33.037 | 0.000 | 10.973 | 12.357 | 1.216e+04 | 388.977 | 31.271 | 0.000 | 1.14e+04 | 1.29e+04 | 5165.1579 | 123.041 | 41.979 | 0.000 | 4924.001 | 5406.314 | 6959.5602 | 196.210 | 35.470 | 0.000 | 6574.996 | 7344.124 | 290.6379 | 7.820 | 37.167 | 0.000 | 275.312 | 305.964 | 1.271e+04 | 463.648 | 27.421 | 0.000 | 1.18e+04 | 1.36e+04 | 1.009e+04 | 532.211 | 18.957 | 0.000 | 9045.861 | 1.11e+04 | 0.0846 | 0.037 | 2.315 | 0.021 | 0.013 | 0.156 | 2.0488 | 1.034 | 1.981 | 0.048 | 0.022 | 4.076 | 0.0007 | 6.85e-05 | 9.656 | 0.000 | 0.001 | 0.001 | |
| $\\lambda_c$ | frequency.cycle | 0.3549 | 0.007 | 54.377 | 0.000 | 0.342 | 0.368 | 0.3670 | 0.007 | 55.056 | 0.000 | 0.354 | 0.380 | 0.4027 | 0.024 | 16.702 | 0.000 | 0.355 | 0.450 | 0.3673 | 0.007 | 49.527 | 0.000 | 0.353 | 0.382 | 0.3473 | 0.006 | 54.072 | 0.000 | 0.335 | 0.360 | 0.3254 | 0.005 | 65.489 | 0.000 | 0.316 | 0.335 | 0.3589 | 0.005 | 74.021 | 0.000 | 0.349 | 0.368 | 0.3047 | 0.005 | 57.097 | 0.000 | 0.294 | 0.315 | 0.4124 | 0.009 | 46.494 | 0.000 | 0.395 | 0.430 | 0.5102 | 0.006 | 81.920 | 0.000 | 0.498 | 0.522 | 1.0259 | 0.039 | 26.588 | 0.000 | 0.950 | 1.102 | 0.8665 | 0.069 | 12.477 | 0.000 | 0.730 | 1.003 | 0.2907 | 0.007 | 40.772 | 0.000 | 0.277 | 0.305 | |
| $\\rho$ | damping.cycle | 0.8464 | 0.005 | 173.103 | 0.000 | 0.837 | 0.856 | 0.8637 | 0.004 | 194.453 | 0.000 | 0.855 | 0.872 | 0.8109 | 0.016 | 51.546 | 0.000 | 0.780 | 0.842 | 0.8382 | 0.005 | 177.924 | 0.000 | 0.829 | 0.847 | 0.8692 | 0.004 | 202.716 | 0.000 | 0.861 | 0.878 | 0.8654 | 0.004 | 230.590 | 0.000 | 0.858 | 0.873 | 0.8891 | 0.004 | 226.822 | 0.000 | 0.881 | 0.897 | 0.8834 | 0.004 | 225.678 | 0.000 | 0.876 | 0.891 | 0.8314 | 0.005 | 161.369 | 0.000 | 0.821 | 0.842 | 0.8922 | 0.005 | 169.447 | 0.000 | 0.882 | 0.903 | 0.7890 | 0.038 | 20.866 | 0.000 | 0.715 | 0.863 | 0.7217 | 0.043 | 16.811 | 0.000 | 0.638 | 0.806 | 0.9246 | 0.005 | 173.539 | 0.000 | 0.914 | 0.935 | |
| $2 \\pi / \\lambda_c$ | period.cycle | 17.704101 | NaN | NaN | NaN | NaN | NaN | 17.120396 | NaN | NaN | NaN | NaN | NaN | 15.602645 | NaN | NaN | NaN | NaN | NaN | 17.106412 | NaN | NaN | NaN | NaN | NaN | 18.091521 | NaN | NaN | NaN | NaN | NaN | 19.309113 | NaN | NaN | NaN | NaN | NaN | 17.506785 | NaN | NaN | NaN | NaN | NaN | 20.62089 | NaN | NaN | NaN | NaN | NaN | 15.235658 | NaN | NaN | NaN | NaN | NaN | 12.315142 | NaN | NaN | NaN | NaN | NaN | 6.124559 | NaN | NaN | NaN | NaN | NaN | 7.251224 | NaN | NaN | NaN | NaN | NaN | 21.613985 | NaN | NaN | NaN | NaN | NaN | |
Descriptive Statistics
Cycle subseries description


Monthly description

cross-sectional description
| Date | 2004-03 | 2004-04 | 2004-05 | 2004-06 | 2004-07 | 2004-08 | 2004-09 | 2004-10 | 2004-11 | 2004-12 | 2005-01 | 2005-02 | 2005-03 | 2005-04 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CO(GT) | count | 510.0000 | 720.0000 | 744.0000 | 720.0000 | 744.0000 | 744.0000 | 720.0000 | 744.0000 | 720.0000 | 744.0000 | 744.0000 | 672.0000 | 744.0000 | 87.0000 |
| min | 0.3000 | 0.3000 | 0.1000 | 0.1000 | 0.1000 | 0.1000 | 0.2000 | 0.4000 | 0.1000 | 0.2000 | 0.1000 | 0.3000 | 0.1000 | 0.3000 | |
| max | 8.1000 | 7.3000 | 6.5000 | 6.4000 | 5.3000 | 3.5000 | 7.5000 | 9.5000 | 11.9000 | 9.9000 | 8.7000 | 8.4000 | 7.5000 | 5.0000 | |
| mean | 2.2483 | 2.2404 | 1.8668 | 1.8688 | 1.7521 | 1.2496 | 2.1084 | 2.6144 | 2.6496 | 2.7188 | 2.0857 | 2.0822 | 2.0136 | 1.1936 | |
| std | 1.3503 | 1.2551 | 1.1217 | 1.1032 | 0.9886 | 0.4999 | 1.3109 | 1.3611 | 1.8917 | 1.6800 | 1.3695 | 1.3120 | 1.3764 | 0.8170 | |
| PT08.S1(CO) | count | 510.0000 | 720.0000 | 744.0000 | 720.0000 | 744.0000 | 744.0000 | 720.0000 | 744.0000 | 720.0000 | 744.0000 | 744.0000 | 672.0000 | 744.0000 | 87.0000 |
| min | 818.0000 | 753.0000 | 736.0000 | 708.0000 | 723.0000 | 711.0000 | 718.0000 | 704.0000 | 647.0000 | 691.0000 | 744.0000 | 755.0000 | 715.0000 | 811.0000 | |
| max | 2040.0000 | 1875.0000 | 1763.0000 | 1506.0000 | 1626.0000 | 1393.0000 | 1728.0000 | 1908.0000 | 2008.0000 | 1881.0000 | 1835.0000 | 1846.0000 | 1818.0000 | 1446.0000 | |
| mean | 1222.8196 | 1161.7003 | 1075.4309 | 1019.2030 | 1046.3647 | 979.5625 | 1079.9986 | 1184.9679 | 1132.1486 | 1084.4706 | 1109.0407 | 1083.8458 | 1145.3640 | 953.7126 | |
| std | 241.0385 | 228.0895 | 194.4596 | 159.0491 | 158.3249 | 117.1149 | 198.3246 | 242.9786 | 273.0584 | 213.1563 | 215.5116 | 185.2794 | 197.1283 | 125.4414 | |
| NMHC(GT) | count | 510.0000 | 720.0000 | 1.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| min | 7.0000 | 9.0000 | 275.0000 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | |
| max | 797.0000 | 1189.0000 | 275.0000 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | |
| mean | 170.5805 | 252.4566 | 275.0000 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | |
| std | 141.6348 | 202.9348 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | |
| C6H6(GT) | count | 510.0000 | 720.0000 | 744.0000 | 720.0000 | 744.0000 | 744.0000 | 720.0000 | 744.0000 | 720.0000 | 744.0000 | 744.0000 | 672.0000 | 744.0000 | 87.0000 |
| min | 0.6000 | 0.5000 | 0.5000 | 0.5000 | 1.3000 | 1.0000 | 0.9000 | 0.5000 | 0.2000 | 0.4000 | 0.1000 | 0.2000 | 0.2000 | 0.8000 | |
| max | 39.2000 | 40.3000 | 40.2000 | 36.9000 | 37.3000 | 30.7000 | 41.2000 | 52.1000 | 63.7000 | 50.8000 | 43.0000 | 33.9000 | 35.5000 | 22.4000 | |
| mean | 9.9327 | 10.3847 | 10.1865 | 10.5798 | 10.5968 | 6.9949 | 11.5172 | 13.5276 | 12.5118 | 9.5040 | 9.0246 | 7.4913 | 8.7322 | 4.2851 | |
| std | 7.0605 | 7.4249 | 6.6894 | 6.4530 | 6.2274 | 3.7573 | 7.9367 | 9.1119 | 9.8386 | 7.0804 | 6.9725 | 5.4261 | 6.6770 | 3.7984 | |
| PT08.S2(NMHC) | count | 510.0000 | 720.0000 | 744.0000 | 720.0000 | 744.0000 | 744.0000 | 720.0000 | 744.0000 | 720.0000 | 744.0000 | 744.0000 | 672.0000 | 744.0000 | 87.0000 |
| min | 457.0000 | 448.0000 | 437.0000 | 437.0000 | 527.0000 | 494.0000 | 486.0000 | 444.0000 | 397.0000 | 426.0000 | 383.0000 | 402.0000 | 387.0000 | 473.0000 | |
| max | 1754.0000 | 1776.0000 | 1776.0000 | 1705.0000 | 1713.0000 | 1567.0000 | 1795.0000 | 2007.0000 | 2214.0000 | 1983.0000 | 1831.0000 | 1639.0000 | 1675.0000 | 1362.0000 | |
| mean | 935.6431 | 948.4665 | 949.4093 | 964.9694 | 971.3774 | 837.5784 | 991.7029 | 1058.5105 | 1011.2014 | 913.4517 | 890.5241 | 839.7649 | 887.9157 | 698.7356 | |
| std | 258.9974 | 269.3944 | 243.8496 | 237.7542 | 224.4253 | 155.9563 | 272.4993 | 294.2151 | 327.3430 | 257.8404 | 268.6751 | 218.1439 | 253.3175 | 173.3043 | |
| NOx(GT) | count | 510.0000 | 720.0000 | 744.0000 | 720.0000 | 744.0000 | 744.0000 | 720.0000 | 744.0000 | 720.0000 | 744.0000 | 744.0000 | 672.0000 | 744.0000 | 87.0000 |
| min | 12.0000 | 10.0000 | 4.0000 | 9.0000 | 10.0000 | 2.0000 | 19.0000 | 33.0000 | 13.0000 | 31.0000 | 17.0000 | 25.0000 | 29.0000 | 30.0000 | |
| max | 478.0000 | 378.0000 | 383.0000 | 441.0000 | 427.0000 | 282.0000 | 891.0000 | 952.0000 | 1389.0000 | 1479.0000 | 1230.0000 | 1227.0000 | 959.0000 | 594.0000 | |
| mean | 139.7090 | 134.7195 | 118.1636 | 118.2016 | 123.3433 | 73.3715 | 267.1590 | 322.0476 | 412.2313 | 405.8573 | 341.4330 | 307.9954 | 306.8580 | 175.5747 | |
| std | 82.8739 | 70.7231 | 65.1873 | 77.7787 | 81.1308 | 40.6644 | 149.5296 | 154.6412 | 284.8950 | 248.9821 | 232.9381 | 207.1190 | 188.4357 | 109.4848 | |
| PT08.S3(NOx) | count | 510.0000 | 720.0000 | 744.0000 | 720.0000 | 744.0000 | 744.0000 | 720.0000 | 744.0000 | 720.0000 | 744.0000 | 744.0000 | 672.0000 | 744.0000 | 87.0000 |
| min | 537.0000 | 461.0000 | 452.0000 | 459.0000 | 410.0000 | 416.0000 | 388.0000 | 325.0000 | 322.0000 | 334.0000 | 355.0000 | 383.0000 | 330.0000 | 415.0000 | |
| max | 1935.0000 | 1923.0000 | 1904.0000 | 1941.0000 | 1274.0000 | 1398.0000 | 1531.0000 | 1590.0000 | 2121.0000 | 2683.0000 | 1881.0000 | 1665.0000 | 1804.0000 | 1257.0000 | |
| mean | 1029.1725 | 934.3643 | 947.8272 | 898.8842 | 804.8553 | 833.6421 | 810.5808 | 688.0169 | 789.8806 | 878.6447 | 789.3091 | 788.2502 | 709.2681 | 899.4713 | |
| std | 268.1309 | 253.0501 | 233.0840 | 214.4089 | 163.6861 | 155.5296 | 211.5278 | 203.7557 | 315.4736 | 331.5311 | 258.1564 | 203.3980 | 216.4239 | 172.8037 | |
| NO2(GT) | count | 510.0000 | 720.0000 | 744.0000 | 720.0000 | 744.0000 | 744.0000 | 720.0000 | 744.0000 | 720.0000 | 744.0000 | 744.0000 | 672.0000 | 744.0000 | 87.0000 |
| min | 20.0000 | 17.0000 | 8.0000 | 16.0000 | 17.0000 | 5.0000 | 2.0000 | 29.0000 | 13.0000 | 27.0000 | 17.0000 | 25.0000 | 27.0000 | 29.0000 | |
| max | 194.0000 | 196.0000 | 179.0000 | 226.0000 | 233.0000 | 185.0000 | 225.0000 | 194.0000 | 288.0000 | 269.0000 | 285.0000 | 340.0000 | 248.0000 | 190.0000 | |
| mean | 100.8606 | 93.9107 | 90.7028 | 92.1076 | 102.1456 | 67.8495 | 103.4455 | 86.2438 | 119.9897 | 122.1628 | 132.6206 | 158.1743 | 136.3770 | 108.9310 | |
| std | 32.5799 | 29.2103 | 30.2780 | 38.7665 | 40.3100 | 24.8443 | 40.9144 | 27.3533 | 53.8030 | 40.3027 | 46.1709 | 57.9836 | 45.5583 | 40.4730 | |
| PT08.S4(NO2) | count | 510.0000 | 720.0000 | 744.0000 | 720.0000 | 744.0000 | 744.0000 | 720.0000 | 744.0000 | 720.0000 | 744.0000 | 744.0000 | 672.0000 | 744.0000 | 87.0000 |
| min | 1050.0000 | 955.0000 | 1048.0000 | 1227.0000 | 1234.0000 | 1084.0000 | 1059.0000 | 1050.0000 | 697.0000 | 682.0000 | 657.0000 | 621.0000 | 551.0000 | 742.0000 | |
| max | 2679.0000 | 2684.0000 | 2667.0000 | 2746.0000 | 2662.0000 | 2404.0000 | 2555.0000 | 2775.0000 | 2643.0000 | 2405.0000 | 2083.0000 | 1870.0000 | 2147.0000 | 1777.0000 | |
| mean | 1572.0725 | 1609.0432 | 1599.5351 | 1719.4459 | 1643.7175 | 1577.9504 | 1544.7438 | 1636.9817 | 1372.2347 | 1213.9987 | 1151.4542 | 1053.9850 | 1225.3955 | 976.4598 | |
| std | 280.5254 | 287.1709 | 261.0709 | 244.9339 | 226.3288 | 181.8614 | 286.1289 | 302.1502 | 355.8909 | 263.3822 | 271.0564 | 207.4259 | 275.9075 | 169.7447 | |
| PT08.S5(O3) | count | 510.0000 | 720.0000 | 744.0000 | 720.0000 | 744.0000 | 744.0000 | 720.0000 | 744.0000 | 720.0000 | 744.0000 | 744.0000 | 672.0000 | 744.0000 | 87.0000 |
| min | 341.0000 | 263.0000 | 301.0000 | 334.0000 | 386.0000 | 368.0000 | 342.0000 | 392.0000 | 261.0000 | 318.0000 | 253.0000 | 252.0000 | 221.0000 | 322.0000 | |
| max | 2359.0000 | 2108.0000 | 2054.0000 | 2202.0000 | 2475.0000 | 1607.0000 | 2321.0000 | 2372.0000 | 2522.0000 | 2523.0000 | 2331.0000 | 2494.0000 | 2159.0000 | 1729.0000 | |
| mean | 1027.4941 | 1002.7288 | 927.2517 | 935.6565 | 994.4457 | 775.6035 | 1034.3404 | 1163.9053 | 1171.3542 | 1080.6730 | 1096.5487 | 1028.9280 | 1083.8291 | 646.8506 | |
| std | 381.6004 | 376.9071 | 324.1343 | 306.4241 | 352.3788 | 221.8787 | 355.5132 | 377.5972 | 469.6605 | 413.4739 | 445.1649 | 416.3604 | 408.9590 | 305.9041 | |
| T | count | 510.0000 | 720.0000 | 744.0000 | 720.0000 | 744.0000 | 744.0000 | 720.0000 | 744.0000 | 720.0000 | 744.0000 | 744.0000 | 672.0000 | 744.0000 | 87.0000 |
| min | 6.1000 | 7.4000 | 7.4000 | 16.5000 | 16.2000 | 18.6000 | 11.9000 | 12.6000 | 1.3000 | 1.2000 | 1.0000 | 0.3000 | -1.9000 | 8.9000 | |
| max | 29.3000 | 31.3000 | 32.8000 | 42.2000 | 44.6000 | 40.5000 | 40.7000 | 30.8000 | 26.8000 | 20.3000 | 16.9000 | 19.9000 | 28.8000 | 30.0000 | |
| mean | 14.3908 | 16.8004 | 20.2230 | 26.4517 | 29.4131 | 29.1182 | 24.5979 | 20.5012 | 13.4854 | 11.1088 | 8.2079 | 7.0960 | 13.2206 | 16.6609 | |
| std | 4.4475 | 4.7505 | 5.1867 | 5.8866 | 6.1716 | 5.1330 | 5.4366 | 3.5880 | 4.9904 | 3.4512 | 3.7218 | 3.3541 | 6.3927 | 5.4252 | |
| RH | count | 510.0000 | 720.0000 | 744.0000 | 720.0000 | 744.0000 | 744.0000 | 720.0000 | 744.0000 | 720.0000 | 744.0000 | 744.0000 | 672.0000 | 744.0000 | 87.0000 |
| min | 14.9000 | 16.2000 | 9.2000 | 11.9000 | 9.6000 | 10.0000 | 13.8000 | 31.8000 | 14.0000 | 16.3000 | 17.2000 | 18.0000 | 13.5000 | 9.9000 | |
| max | 83.2000 | 82.4000 | 85.2000 | 82.3000 | 69.3000 | 81.8000 | 87.2000 | 86.5000 | 87.1000 | 88.7000 | 86.6000 | 86.6000 | 84.0000 | 63.1000 | |
| mean | 50.1708 | 50.5945 | 43.6366 | 39.7443 | 33.0613 | 42.7673 | 43.9763 | 61.8939 | 59.2749 | 57.6148 | 56.4503 | 51.8323 | 50.8980 | 35.4828 | |
| std | 14.2653 | 15.9084 | 16.6971 | 13.9907 | 12.7009 | 14.4153 | 15.0220 | 11.2454 | 16.0088 | 16.4268 | 13.8457 | 16.4541 | 16.6005 | 13.1440 | |
| AH | count | 510.0000 | 720.0000 | 744.0000 | 720.0000 | 744.0000 | 744.0000 | 720.0000 | 744.0000 | 720.0000 | 744.0000 | 744.0000 | 672.0000 | 744.0000 | 87.0000 |
| min | 0.4023 | 0.4868 | 0.3754 | 0.7502 | 0.7158 | 0.6591 | 0.6514 | 0.9107 | 0.1988 | 0.2749 | 0.2477 | 0.2269 | 0.1847 | 0.3866 | |
| max | 1.0945 | 1.4852 | 1.6296 | 1.9390 | 2.0042 | 2.1806 | 2.2310 | 2.0224 | 1.9800 | 1.3713 | 1.0567 | 1.0859 | 1.3930 | 0.8642 | |
| mean | 0.7894 | 0.9196 | 0.9572 | 1.2733 | 1.2427 | 1.6153 | 1.2846 | 1.4640 | 0.9400 | 0.7780 | 0.6341 | 0.5153 | 0.7817 | 0.6216 | |
| std | 0.1399 | 0.1699 | 0.1985 | 0.1995 | 0.2434 | 0.2862 | 0.2919 | 0.2195 | 0.3603 | 0.2741 | 0.2185 | 0.1405 | 0.2916 | 0.1164 |

longitudinal description
| Autocorrelation | Heteroskedasticity | Normality | Stationary | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2004-03 | 2004-04 | 2004-05 | 2004-06 | 2004-07 | 2004-08 | 2004-09 | 2004-10 | 2004-11 | 2004-12 | 2005-01 | 2005-02 | 2004-03 | 2004-04 | 2004-05 | 2004-06 | 2004-07 | 2004-08 | 2004-09 | 2004-10 | 2004-11 | 2004-12 | 2005-01 | 2005-02 | 2004-03 | 2004-04 | 2004-05 | 2004-06 | 2004-07 | 2004-08 | 2004-09 | 2004-10 | 2004-11 | 2004-12 | 2005-01 | 2005-02 | 2004-03 | 2004-04 | 2004-05 | 2004-06 | 2004-07 | 2004-08 | 2004-09 | 2004-10 | 2004-11 | 2004-12 | 2005-01 | 2005-02 | |
| CO(GT) | True | True | True | True | True | True | True | True | True | True | True | True | False | False | True | False | True | False | True | True | True | True | True | False | False | False | False | False | False | False | False | False | False | False | False | False | False | False | False | True | True | True | True | True | True | True | False | True |
| PT08.S1(CO) | True | True | True | True | True | True | True | True | True | True | True | True | False | False | False | False | False | False | False | False | False | False | True | False | False | False | False | False | False | False | False | False | False | False | False | False | False | False | True | True | True | False | False | False | False | False | False | False |
| C6H6(GT) | True | True | True | True | True | True | True | True | True | True | True | True | False | False | False | False | False | False | True | False | False | True | True | True | False | False | False | False | False | False | False | False | False | False | False | False | False | False | True | True | True | False | True | True | True | False | False | False |
| PT08.S2(NMHC) | True | True | True | True | True | True | True | True | True | True | True | True | False | False | False | False | False | False | False | False | False | False | True | False | False | False | False | False | False | False | False | False | False | False | False | False | False | False | True | True | True | False | True | True | False | False | False | True |
| NOx(GT) | True | True | True | True | True | True | True | True | True | True | True | True | True | True | True | False | True | False | True | True | True | False | True | True | False | False | False | False | False | False | False | False | False | False | False | False | False | False | False | True | False | True | True | True | False | False | False | False |
| PT08.S3(NOx) | True | True | True | True | True | True | True | True | True | True | True | True | True | False | False | False | False | False | False | False | False | True | True | False | False | False | False | False | False | False | False | False | False | False | False | False | False | False | False | False | True | False | False | False | False | True | False | False |
| NO2(GT) | True | True | True | True | True | True | True | True | True | True | True | True | True | False | False | False | False | False | False | True | True | True | False | False | False | False | True | False | False | False | False | False | False | False | True | False | False | False | True | False | False | True | False | False | False | False | True | False |
| PT08.S4(NO2) | True | True | True | True | True | True | True | True | True | True | True | True | True | False | False | False | False | False | False | False | False | True | True | False | False | True | False | False | False | False | False | False | False | False | False | False | False | False | False | True | True | False | False | False | False | False | False | False |
| PT08.S5(O3) | True | True | True | True | True | True | True | True | True | True | True | True | False | False | False | False | False | False | False | False | True | False | True | True | False | False | False | False | False | False | False | False | False | False | False | False | False | False | True | True | True | True | False | False | False | False | False | False |
| T | True | True | True | True | True | True | True | True | True | True | True | True | True | False | True | True | False | False | True | False | True | True | True | True | True | False | False | False | False | False | False | False | False | True | False | False | False | False | False | False | False | False | False | False | False | False | False | False |
| RH | True | True | True | True | True | True | True | True | True | True | True | True | False | True | True | True | False | True | True | True | False | True | True | True | False | False | False | False | False | False | False | False | False | False | False | False | True | False | False | False | False | False | False | False | False | False | False | False |
| AH | True | True | True | True | True | True | True | True | True | True | True | True | True | False | True | False | False | True | False | False | True | True | True | True | True | True | False | True | False | False | False | False | False | False | False | False | False | True | False | True | False | False | True | True | False | False | False | False |
| NMHC(GT) | True | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | False | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | False | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | True | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
Weekly description

cross-sectional description
| Date | 2004-03-14 | 2004-03-21 | 2004-03-28 | 2004-04-04 | 2004-04-11 | 2004-04-18 | 2004-04-25 | 2004-05-02 | 2004-05-09 | 2004-05-16 | 2004-05-23 | 2004-05-30 | 2004-06-06 | 2004-06-13 | 2004-06-20 | 2004-06-27 | 2004-07-04 | 2004-07-11 | 2004-07-18 | 2004-07-25 | 2004-08-01 | 2004-08-08 | 2004-08-15 | 2004-08-22 | 2004-08-29 | 2004-09-05 | 2004-09-12 | 2004-09-19 | 2004-09-26 | 2004-10-03 | 2004-10-10 | 2004-10-17 | 2004-10-24 | 2004-10-31 | 2004-11-07 | 2004-11-14 | 2004-11-21 | 2004-11-28 | 2004-12-05 | 2004-12-12 | 2004-12-19 | 2004-12-26 | 2005-01-02 | 2005-01-09 | 2005-01-16 | 2005-01-23 | 2005-01-30 | 2005-02-06 | 2005-02-13 | 2005-02-20 | 2005-02-27 | 2005-03-06 | 2005-03-13 | 2005-03-20 | 2005-03-27 | 2005-04-03 | 2005-04-10 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CO(GT) | count | 102.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 15.0000 |
| min | 0.6000 | 0.5000 | 0.3000 | 0.5000 | 0.3000 | 0.3000 | 0.3000 | 0.3000 | 0.4000 | 0.2000 | 0.2000 | 0.1000 | 0.1000 | 0.3000 | 0.3000 | 0.2000 | 0.3000 | 0.2000 | 0.1000 | 0.1000 | 0.4000 | 0.1000 | 0.4000 | 0.3000 | 0.4000 | 0.5000 | 0.2000 | 0.3000 | 0.3000 | 0.4000 | 0.7000 | 0.4000 | 0.7000 | 0.4000 | 0.4000 | 0.2000 | 0.1000 | 0.1000 | 0.6000 | 0.3000 | 0.6000 | 0.2000 | 0.4000 | 0.5000 | 0.4000 | 0.1000 | 0.1000 | 0.3000 | 0.4000 | 0.3000 | 0.4000 | 0.2000 | 0.1000 | 0.1000 | 0.1000 | 0.3000 | 0.4000 | |
| max | 6.9000 | 8.1000 | 5.2000 | 6.2000 | 6.3000 | 7.3000 | 5.6000 | 7.2000 | 5.4000 | 6.3000 | 6.5000 | 4.0000 | 4.8000 | 6.4000 | 5.0000 | 5.0000 | 5.7000 | 5.3000 | 5.0000 | 4.4000 | 3.4000 | 3.0000 | 2.9000 | 2.5000 | 3.5000 | 5.8000 | 5.4000 | 6.7000 | 7.3000 | 7.5000 | 7.0000 | 4.8000 | 8.4000 | 9.5000 | 9.2000 | 6.8000 | 10.2000 | 11.9000 | 9.4000 | 6.3000 | 9.9000 | 9.1000 | 4.7000 | 5.9000 | 5.9000 | 8.7000 | 3.0000 | 6.8000 | 8.4000 | 5.6000 | 6.3000 | 6.1000 | 6.0000 | 7.4000 | 7.5000 | 5.1000 | 5.0000 | |
| mean | 2.4506 | 2.6537 | 1.8927 | 2.1650 | 2.0068 | 2.3141 | 2.1462 | 2.2720 | 2.0241 | 2.1638 | 1.9963 | 1.3697 | 1.6205 | 1.9865 | 1.7730 | 1.8568 | 1.9047 | 1.8246 | 1.8738 | 1.6795 | 1.5913 | 1.2884 | 1.1648 | 1.2025 | 1.3420 | 1.7837 | 1.9728 | 1.9478 | 2.2826 | 2.5729 | 2.4509 | 2.4985 | 2.7158 | 2.7852 | 2.7045 | 1.9112 | 2.5132 | 3.4701 | 2.7870 | 2.0686 | 3.1863 | 3.0270 | 2.3226 | 2.3946 | 2.5722 | 2.2430 | 1.0632 | 2.2738 | 2.5226 | 1.7012 | 2.0538 | 1.7202 | 1.7614 | 2.2780 | 2.2935 | 1.4516 | 1.9848 | |
| std | 1.3316 | 1.6240 | 0.9951 | 1.1587 | 1.1074 | 1.4008 | 1.0486 | 1.3669 | 1.0485 | 1.2524 | 1.2566 | 0.7866 | 0.8287 | 1.3338 | 1.0076 | 1.0246 | 1.1261 | 1.2067 | 1.1663 | 0.8714 | 0.6105 | 0.5345 | 0.5129 | 0.3892 | 0.5119 | 1.0108 | 0.9894 | 1.1139 | 1.5753 | 1.4534 | 1.1765 | 1.0516 | 1.4513 | 1.7545 | 1.8198 | 1.3615 | 1.8546 | 2.1823 | 1.7779 | 1.0741 | 1.9423 | 1.9507 | 0.9234 | 1.2384 | 1.3097 | 1.6201 | 0.5877 | 1.5138 | 1.5621 | 0.9233 | 1.1288 | 1.1677 | 1.3583 | 1.4975 | 1.4849 | 0.9246 | 1.4878 | |
| PT08.S1(CO) | count | 102.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 15.0000 |
| min | 831.0000 | 869.0000 | 834.0000 | 818.0000 | 797.0000 | 753.0000 | 763.0000 | 767.0000 | 743.0000 | 787.0000 | 771.0000 | 738.0000 | 729.0000 | 726.0000 | 708.0000 | 719.0000 | 809.0000 | 775.0000 | 723.0000 | 834.0000 | 742.0000 | 788.0000 | 738.0000 | 711.0000 | 749.0000 | 784.0000 | 806.0000 | 752.0000 | 718.0000 | 741.0000 | 820.0000 | 704.0000 | 760.0000 | 791.0000 | 752.0000 | 647.0000 | 667.0000 | 814.0000 | 797.0000 | 709.0000 | 699.0000 | 691.0000 | 744.0000 | 862.0000 | 819.0000 | 795.0000 | 744.0000 | 787.0000 | 844.0000 | 802.0000 | 844.0000 | 715.0000 | 841.0000 | 911.0000 | 878.0000 | 811.0000 | 864.0000 | |
| max | 1898.0000 | 2040.0000 | 1597.0000 | 1722.0000 | 1757.0000 | 1875.0000 | 1747.0000 | 1771.0000 | 1597.0000 | 1754.0000 | 1763.0000 | 1485.0000 | 1501.0000 | 1496.0000 | 1440.0000 | 1332.0000 | 1529.0000 | 1611.0000 | 1339.0000 | 1626.0000 | 1380.0000 | 1381.0000 | 1268.0000 | 1175.0000 | 1314.0000 | 1488.0000 | 1506.0000 | 1571.0000 | 1600.0000 | 1728.0000 | 1840.0000 | 1464.0000 | 1884.0000 | 1908.0000 | 1915.0000 | 1578.0000 | 1802.0000 | 2008.0000 | 1816.0000 | 1604.0000 | 1881.0000 | 1739.0000 | 1472.0000 | 1741.0000 | 1526.0000 | 1835.0000 | 1198.6590 | 1713.0000 | 1846.0000 | 1464.0000 | 1527.0000 | 1567.0000 | 1573.0000 | 1710.0000 | 1818.0000 | 1531.0000 | 1446.0000 | |
| mean | 1305.1176 | 1336.1369 | 1103.5774 | 1199.0624 | 1116.2187 | 1139.5357 | 1142.7857 | 1168.7381 | 1058.7381 | 1122.3155 | 1154.8452 | 975.2656 | 984.5714 | 1027.4702 | 1002.6683 | 1008.7851 | 1084.8274 | 1060.1131 | 1000.8333 | 1118.4762 | 997.1924 | 1025.6429 | 973.1601 | 937.7024 | 957.0334 | 1059.4762 | 1078.7620 | 1079.4881 | 1076.9167 | 1160.8818 | 1258.4048 | 1000.2321 | 1274.0357 | 1193.4405 | 1188.2500 | 970.2202 | 1045.9583 | 1315.3155 | 1170.6369 | 1041.0536 | 1099.0182 | 1126.2505 | 1013.8838 | 1165.2902 | 1205.6488 | 1165.3393 | 904.3871 | 1109.8631 | 1147.0618 | 1046.5119 | 1067.7024 | 1003.2619 | 1117.3204 | 1204.7976 | 1243.6786 | 1018.9643 | 1090.5333 | |
| std | 218.3815 | 259.8206 | 160.3252 | 209.4062 | 195.4378 | 226.5464 | 239.8161 | 238.4217 | 179.8965 | 206.6689 | 204.6472 | 148.7877 | 150.7783 | 175.5138 | 138.3322 | 138.5456 | 161.8965 | 184.4432 | 128.1401 | 165.2033 | 120.4266 | 123.8964 | 113.4518 | 99.5979 | 97.0059 | 151.2495 | 152.0301 | 200.5149 | 232.9801 | 225.1351 | 237.1526 | 165.1213 | 242.0687 | 238.7695 | 269.0931 | 197.0507 | 242.8034 | 262.4005 | 228.1977 | 168.8166 | 192.2486 | 252.5993 | 148.7457 | 174.6899 | 198.3606 | 238.9035 | 100.4423 | 201.6630 | 224.9558 | 123.3061 | 149.9145 | 186.6587 | 173.2835 | 180.8384 | 200.6301 | 151.1370 | 192.3185 | |
| NMHC(GT) | count | 102.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 121.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| min | 7.0000 | 11.0000 | 19.0000 | 18.0000 | 30.0000 | 9.0000 | 20.0000 | 17.0000 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | |
| max | 488.0000 | 685.0000 | 797.0000 | 1042.0000 | 1084.0000 | 1189.0000 | 899.0000 | 1129.0000 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | |
| mean | 127.4811 | 185.7475 | 170.7941 | 250.8988 | 215.0660 | 260.3191 | 248.4291 | 267.6529 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | |
| std | 101.5640 | 128.1449 | 145.6729 | 205.3381 | 190.0289 | 215.2524 | 178.4855 | 224.4483 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | |
| C6H6(GT) | count | 102.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 15.0000 |
| min | 1.0000 | 1.3000 | 0.6000 | 0.8000 | 0.7000 | 0.5000 | 0.7000 | 0.6000 | 1.0000 | 1.0000 | 1.2000 | 0.5000 | 1.1000 | 1.2000 | 0.5000 | 1.2000 | 2.6000 | 2.2000 | 1.4000 | 1.3000 | 1.8000 | 1.6000 | 1.5000 | 1.0000 | 1.7000 | 1.7000 | 1.0000 | 1.2000 | 0.9000 | 1.9000 | 2.5000 | 0.5000 | 1.6000 | 1.2000 | 1.2000 | 0.2000 | 0.4000 | 1.5000 | 1.2000 | 0.5000 | 1.8000 | 0.5000 | 0.4000 | 1.4000 | 1.3000 | 0.7000 | 0.1000 | 0.3000 | 0.6000 | 0.5000 | 0.5000 | 0.2000 | 0.5000 | 1.1000 | 1.2000 | 0.8000 | 0.8000 | |
| max | 32.6000 | 39.2000 | 24.6000 | 31.9000 | 33.7000 | 33.7000 | 40.3000 | 36.2000 | 26.5000 | 37.4000 | 38.9000 | 40.2000 | 27.9000 | 36.9000 | 30.2000 | 30.2000 | 33.5000 | 31.0000 | 30.4000 | 37.3000 | 26.7000 | 26.0000 | 18.7000 | 16.0000 | 21.0000 | 35.2000 | 34.5000 | 35.9000 | 41.2000 | 39.3000 | 44.7000 | 25.6000 | 43.7000 | 52.1000 | 48.2000 | 32.3000 | 47.7000 | 63.7000 | 43.9000 | 26.4000 | 50.8000 | 36.3000 | 23.4000 | 25.3000 | 28.4000 | 43.0000 | 13.5298 | 32.4000 | 26.8000 | 33.9000 | 27.9000 | 28.0000 | 27.0000 | 33.5000 | 35.5000 | 26.3000 | 22.4000 | |
| mean | 10.0647 | 12.6423 | 8.1214 | 9.4592 | 8.8895 | 8.9256 | 11.5554 | 11.2310 | 9.2524 | 11.7476 | 11.0226 | 9.2386 | 9.1494 | 11.5696 | 10.3668 | 10.0806 | 11.2304 | 11.2768 | 10.6315 | 11.1583 | 9.2114 | 8.2232 | 6.2918 | 5.7815 | 6.7784 | 10.9036 | 10.1461 | 11.1196 | 12.0655 | 13.1793 | 14.8548 | 8.6893 | 16.6601 | 14.7214 | 14.8494 | 8.1185 | 10.5929 | 16.8435 | 11.8804 | 8.6220 | 10.9322 | 9.9802 | 5.8564 | 9.3620 | 11.9423 | 11.3595 | 3.7707 | 9.2732 | 8.3489 | 6.5500 | 6.9833 | 6.1190 | 8.2978 | 9.9107 | 10.5446 | 5.6411 | 8.4400 | |
| std | 6.3279 | 8.5044 | 5.3163 | 6.2826 | 5.9616 | 6.8694 | 8.6339 | 7.4288 | 5.8697 | 7.6093 | 7.0110 | 6.4681 | 5.4588 | 7.7917 | 5.8662 | 5.6852 | 6.2450 | 6.9584 | 6.2690 | 6.8935 | 4.9555 | 4.2751 | 3.1604 | 2.5729 | 2.8600 | 6.9386 | 6.0470 | 7.3427 | 9.1214 | 8.6285 | 9.7035 | 5.9828 | 9.2601 | 9.8026 | 10.6848 | 6.8635 | 8.0060 | 11.1612 | 8.9023 | 5.3818 | 7.3049 | 7.0345 | 3.7798 | 5.2468 | 6.9954 | 8.5056 | 3.0201 | 7.3093 | 4.8841 | 4.6467 | 4.8940 | 5.1732 | 6.2447 | 7.1828 | 7.5829 | 4.6747 | 6.8651 | |
| PT08.S2(NMHC) | count | 102.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 15.0000 |
| min | 501.0000 | 525.0000 | 457.0000 | 477.0000 | 464.0000 | 448.0000 | 471.0000 | 454.0000 | 502.0000 | 501.0000 | 521.0000 | 437.0000 | 505.0000 | 520.0000 | 437.0000 | 522.0000 | 624.0000 | 602.0000 | 540.0000 | 527.0000 | 569.0000 | 553.0000 | 544.0000 | 494.0000 | 559.0000 | 565.0000 | 501.0000 | 521.0000 | 486.0000 | 577.0000 | 620.0000 | 444.0000 | 555.0000 | 519.0000 | 516.0000 | 397.0000 | 426.0000 | 548.0000 | 518.0000 | 437.0000 | 567.0000 | 437.0000 | 426.0000 | 540.0000 | 532.0000 | 470.0000 | 383.0000 | 415.0000 | 453.0000 | 437.0000 | 440.0000 | 387.0000 | 443.0000 | 508.0000 | 517.0000 | 473.0000 | 478.0000 | |
| max | 1610.0000 | 1754.0000 | 1418.0000 | 1595.0000 | 1636.0000 | 1637.0000 | 1776.0000 | 1690.0000 | 1466.0000 | 1716.0000 | 1748.0000 | 1776.0000 | 1501.0000 | 1705.0000 | 1556.0000 | 1556.0000 | 1631.0000 | 1575.0000 | 1561.0000 | 1713.0000 | 1472.0000 | 1454.0000 | 1259.0000 | 1181.0000 | 1325.0000 | 1669.0000 | 1654.0000 | 1684.0000 | 1795.0000 | 1756.0000 | 1866.0000 | 1444.0000 | 1846.0000 | 2007.0000 | 1935.0000 | 1605.0000 | 1924.0000 | 2214.0000 | 1851.0000 | 1465.0000 | 1983.0000 | 1692.0000 | 1389.0000 | 1438.0000 | 1512.0000 | 1831.0000 | 1044.2834 | 1605.0000 | 1474.0000 | 1639.0000 | 1502.0000 | 1502.0000 | 1479.0000 | 1632.0000 | 1675.0000 | 1461.0000 | 1362.0000 | |
| mean | 947.0784 | 1030.4405 | 872.2619 | 921.8828 | 895.9199 | 893.6488 | 984.6190 | 984.6488 | 917.2976 | 1004.4524 | 980.1964 | 909.7589 | 917.7917 | 992.3512 | 953.8116 | 953.6227 | 996.2976 | 991.0714 | 972.6786 | 986.5238 | 926.0643 | 889.0774 | 809.8535 | 789.8274 | 828.0534 | 978.2083 | 950.0481 | 983.6726 | 1000.3869 | 1049.8262 | 1101.2619 | 891.8512 | 1165.1012 | 1096.7262 | 1092.1190 | 850.7381 | 950.8810 | 1159.6071 | 998.6190 | 893.7440 | 957.8152 | 933.9233 | 780.2711 | 913.4105 | 1013.5357 | 977.7976 | 666.0265 | 905.8929 | 867.2024 | 808.6548 | 824.8929 | 776.1607 | 873.2757 | 933.7202 | 957.6131 | 762.9702 | 862.2667 | |
| std | 238.5672 | 284.4188 | 220.0662 | 238.9947 | 227.1439 | 262.0269 | 302.1259 | 263.2765 | 228.5458 | 261.8931 | 252.3311 | 240.0438 | 211.7219 | 279.1132 | 225.1439 | 214.4003 | 220.4933 | 247.2923 | 225.4909 | 247.1389 | 188.4348 | 172.6679 | 140.3554 | 122.0652 | 124.9944 | 243.8394 | 221.6805 | 254.0970 | 312.2914 | 282.3680 | 303.8823 | 235.3236 | 282.4253 | 303.9625 | 332.5951 | 281.1881 | 291.5608 | 325.3868 | 297.9177 | 219.2484 | 247.3834 | 264.5793 | 173.1016 | 205.0003 | 253.5595 | 300.2219 | 162.8270 | 269.1022 | 199.5360 | 195.6405 | 208.5739 | 228.9093 | 241.1039 | 262.4726 | 266.7845 | 200.0170 | 295.2310 | |
| NOx(GT) | count | 102.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 15.0000 |
| min | 16.0000 | 18.0000 | 12.0000 | 15.0000 | 12.0000 | 16.0000 | 14.0000 | 10.0000 | 14.0000 | 21.0000 | 14.0000 | 13.0000 | 4.0000 | 19.0000 | 9.0000 | 15.0000 | 24.0000 | 10.0000 | 13.0000 | 10.0000 | 14.0000 | 2.0000 | 6.0000 | 11.0000 | 12.0000 | 35.8227 | 49.0000 | 32.0000 | 19.0000 | 60.0000 | 50.0000 | 33.0000 | 94.0000 | 43.0000 | 44.0000 | 13.0000 | 24.0000 | 60.0000 | 47.0000 | 31.0000 | 83.0000 | 41.0000 | 50.0000 | 54.0000 | 64.0000 | 41.0000 | 17.0000 | 39.0000 | 51.0000 | 28.0000 | 38.0000 | 25.0000 | 35.0000 | 68.0000 | 73.0000 | 30.0000 | 52.0000 | |
| max | 383.0000 | 478.0000 | 275.0000 | 378.0000 | 329.0000 | 352.0000 | 358.0000 | 368.0000 | 320.0000 | 383.0000 | 346.0000 | 289.0000 | 248.0000 | 441.0000 | 311.0000 | 387.0000 | 350.0000 | 392.0000 | 427.0000 | 395.0000 | 297.0000 | 282.0000 | 188.0000 | 190.0000 | 216.0000 | 487.0000 | 631.0000 | 891.0000 | 803.0000 | 799.0000 | 693.0000 | 565.0000 | 895.0000 | 952.0000 | 1052.0000 | 1247.0000 | 1290.0000 | 1389.0000 | 1184.0000 | 968.0000 | 1479.0000 | 1220.0000 | 832.0000 | 1130.0000 | 910.0000 | 1230.0000 | 378.5265 | 1017.0000 | 1227.0000 | 854.0000 | 757.0000 | 959.0000 | 900.0000 | 841.0000 | 874.0000 | 673.0000 | 594.0000 | |
| mean | 156.1910 | 166.6443 | 113.6746 | 130.2265 | 124.9728 | 143.2190 | 126.2778 | 129.7824 | 122.7057 | 136.1644 | 120.0673 | 100.1096 | 102.7416 | 134.0854 | 104.5188 | 119.8590 | 123.6311 | 139.3356 | 139.0624 | 116.7468 | 96.2353 | 85.5326 | 58.0815 | 68.3541 | 78.7968 | 210.4062 | 270.7143 | 253.8619 | 253.6453 | 321.3780 | 291.0993 | 308.7129 | 321.3980 | 366.8319 | 343.1987 | 314.3488 | 405.8784 | 600.5922 | 420.3260 | 307.5024 | 529.7898 | 386.1041 | 345.0517 | 360.6073 | 413.0174 | 425.1157 | 167.2208 | 372.9418 | 359.9968 | 253.9934 | 279.6451 | 268.5826 | 327.5737 | 333.3690 | 316.2500 | 203.3631 | 263.3333 | |
| std | 83.4045 | 96.9249 | 59.4526 | 66.8091 | 67.5853 | 77.7920 | 60.1739 | 75.7332 | 65.1637 | 76.0178 | 68.0286 | 45.4698 | 60.9845 | 93.2354 | 64.6795 | 76.9657 | 76.7388 | 94.7213 | 89.7805 | 78.1290 | 57.0237 | 50.4362 | 30.4865 | 28.9763 | 38.4405 | 116.1754 | 122.5449 | 139.2661 | 179.9484 | 163.2265 | 128.0536 | 118.0611 | 148.1454 | 210.9215 | 214.6588 | 218.4351 | 268.4513 | 335.1495 | 273.6473 | 155.1489 | 321.5270 | 231.8067 | 142.5871 | 206.4254 | 208.7022 | 294.9770 | 85.6651 | 248.6809 | 253.7047 | 145.1205 | 148.4989 | 181.6178 | 203.0508 | 192.4670 | 184.0123 | 122.4991 | 199.5061 | |
| PT08.S3(NOx) | count | 102.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 15.0000 |
| min | 624.0000 | 537.0000 | 652.0000 | 539.0000 | 494.0000 | 482.0000 | 478.0000 | 461.0000 | 518.0000 | 452.0000 | 484.0000 | 632.0000 | 588.0000 | 507.0000 | 517.0000 | 523.0000 | 459.0000 | 472.0000 | 498.0000 | 410.0000 | 470.0000 | 455.0000 | 519.0000 | 580.0000 | 499.0000 | 388.0000 | 529.0000 | 469.0000 | 425.0000 | 411.0000 | 360.0000 | 475.0000 | 348.0000 | 325.0000 | 328.0000 | 429.0000 | 356.0000 | 322.0000 | 341.0000 | 466.0000 | 334.0000 | 402.0000 | 443.0000 | 416.0000 | 414.0000 | 355.0000 | 629.0677 | 383.0000 | 389.0000 | 457.0000 | 411.0000 | 399.0000 | 407.0000 | 346.0000 | 330.0000 | 370.0000 | 415.0000 | |
| max | 1918.0000 | 1620.0000 | 1935.0000 | 1699.0000 | 1826.0000 | 1923.0000 | 1783.0000 | 1826.0000 | 1635.0000 | 1554.0000 | 1566.0000 | 1904.0000 | 1570.0000 | 1534.0000 | 1941.0000 | 1385.0000 | 1185.0000 | 1254.0000 | 1274.0000 | 1242.0000 | 1204.0000 | 1203.0000 | 1281.0000 | 1398.0000 | 1189.0000 | 1327.0000 | 1370.0000 | 1472.0000 | 1531.0000 | 1221.0000 | 1081.0000 | 1590.0000 | 1226.0000 | 1291.0000 | 1370.0000 | 2121.0000 | 1775.0000 | 1207.0000 | 2095.0000 | 2683.0000 | 1731.0000 | 1726.0000 | 1568.0000 | 1110.0000 | 1144.0000 | 1315.0000 | 1881.0000 | 1486.0000 | 1378.0000 | 1491.0000 | 1349.0000 | 1804.0000 | 1490.0000 | 1049.0000 | 1069.0000 | 1257.0000 | 1116.0000 | |
| mean | 1097.7549 | 924.8929 | 1071.5417 | 964.7617 | 977.9307 | 970.8631 | 927.8036 | 892.0595 | 932.1964 | 869.9405 | 883.5714 | 1104.9490 | 993.5298 | 913.8452 | 900.6559 | 863.8894 | 792.2500 | 815.7321 | 826.6786 | 762.3869 | 812.6865 | 783.5417 | 864.4610 | 895.6429 | 822.1624 | 728.7679 | 866.1735 | 828.2321 | 822.2560 | 718.4379 | 644.4345 | 852.8333 | 603.1726 | 656.8512 | 678.6369 | 967.8452 | 849.7560 | 632.5000 | 918.7857 | 944.5357 | 823.9854 | 813.6136 | 871.9893 | 709.1816 | 659.6071 | 732.9167 | 1048.1742 | 766.4583 | 734.9950 | 810.7381 | 796.8929 | 908.1310 | 737.1814 | 645.6429 | 608.3750 | 805.2143 | 745.2667 | |
| std | 294.0592 | 220.5527 | 262.7886 | 236.6762 | 237.8954 | 280.3216 | 262.1432 | 235.2968 | 233.1504 | 218.4521 | 209.6953 | 221.2966 | 197.8279 | 229.0454 | 232.6463 | 169.5926 | 156.1162 | 178.2363 | 155.5282 | 176.7005 | 142.7198 | 147.0559 | 158.1316 | 151.6203 | 123.8031 | 180.7948 | 161.4553 | 203.4579 | 256.5547 | 184.3731 | 170.1049 | 224.0442 | 161.0118 | 186.0237 | 229.8328 | 394.9105 | 290.9927 | 189.2264 | 419.2740 | 413.1304 | 217.8770 | 270.5445 | 194.3199 | 160.1870 | 189.8977 | 230.1775 | 261.2644 | 213.6051 | 184.3915 | 175.1869 | 201.3900 | 291.2265 | 195.7779 | 162.1011 | 153.8856 | 192.7991 | 232.1337 | |
| NO2(GT) | count | 102.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 15.0000 |
| min | 28.0000 | 28.0000 | 20.0000 | 26.0000 | 19.0000 | 25.0000 | 20.0000 | 17.0000 | 22.0000 | 25.0000 | 23.0000 | 19.0000 | 8.0000 | 30.0000 | 16.0000 | 24.0000 | 39.0000 | 19.0000 | 22.0000 | 17.0000 | 21.0000 | 5.0000 | 5.0000 | 12.0000 | 21.0000 | 27.0000 | 41.0000 | 23.0000 | 2.0000 | 45.0000 | 38.0000 | 29.0000 | 42.0000 | 30.0000 | 26.0000 | 13.0000 | 21.0000 | 44.0000 | 38.0000 | 27.0000 | 61.0000 | 37.0000 | 46.0000 | 44.0000 | 54.0000 | 40.0000 | 17.0000 | 37.0000 | 49.0000 | 28.0000 | 36.0000 | 25.0000 | 34.0000 | 57.0000 | 61.0000 | 29.0000 | 43.0000 | |
| max | 173.0000 | 194.0000 | 146.0000 | 168.0000 | 171.0000 | 158.0000 | 196.0000 | 162.0000 | 139.0000 | 164.0000 | 179.0000 | 155.0000 | 161.0000 | 222.0000 | 180.0000 | 166.0000 | 226.0000 | 233.0000 | 206.0000 | 200.0000 | 174.0000 | 185.0000 | 126.0000 | 137.0000 | 129.0000 | 171.0000 | 220.0000 | 208.0000 | 225.0000 | 209.0000 | 128.0000 | 112.0171 | 194.0000 | 187.0000 | 190.0000 | 211.0000 | 288.0000 | 255.0000 | 232.0000 | 188.0000 | 269.0000 | 252.0000 | 191.0000 | 219.0000 | 212.0000 | 272.0000 | 206.0000 | 333.0000 | 340.0000 | 288.0000 | 295.0000 | 240.0000 | 246.0000 | 248.0000 | 232.0000 | 193.0000 | 190.0000 | |
| mean | 109.0975 | 110.2153 | 89.8681 | 96.7054 | 91.7913 | 93.5436 | 91.2034 | 92.2530 | 87.6804 | 96.8520 | 96.3660 | 84.5732 | 75.1668 | 99.8385 | 89.2357 | 93.3222 | 105.1115 | 108.5038 | 103.6710 | 100.0976 | 93.4649 | 79.8183 | 57.6226 | 63.1167 | 71.8033 | 92.6988 | 108.6447 | 95.6611 | 99.3708 | 104.9655 | 80.0986 | 81.8962 | 87.3143 | 94.4870 | 86.7335 | 110.2302 | 147.7827 | 138.0462 | 109.7303 | 105.2719 | 138.9024 | 135.3292 | 119.7700 | 131.1875 | 133.1860 | 150.1594 | 110.9841 | 166.7504 | 171.5197 | 145.4208 | 159.8083 | 134.0945 | 147.3728 | 148.0595 | 124.7560 | 109.6429 | 122.0000 | |
| std | 33.5981 | 35.9756 | 26.2070 | 27.3498 | 28.5154 | 28.0027 | 29.3801 | 31.4027 | 26.6178 | 29.4296 | 34.8529 | 26.2314 | 29.8660 | 43.9144 | 33.8042 | 33.5641 | 42.9618 | 47.1765 | 39.4597 | 41.7895 | 34.8083 | 31.6726 | 20.5955 | 18.7018 | 21.6651 | 30.2429 | 36.3905 | 34.5723 | 52.5089 | 39.7996 | 21.6350 | 22.0239 | 27.8635 | 34.5399 | 36.1457 | 48.3074 | 59.3378 | 50.3425 | 34.0012 | 31.5554 | 48.1052 | 43.4189 | 27.1952 | 37.4464 | 33.9139 | 49.7138 | 46.6959 | 57.6103 | 62.9317 | 52.4980 | 58.0953 | 46.6637 | 47.1658 | 46.6288 | 39.5642 | 35.5136 | 58.7258 | |
| PT08.S4(NO2) | count | 102.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 15.0000 |
| min | 1134.0000 | 1260.0000 | 1158.0000 | 1050.0000 | 1076.0000 | 955.0000 | 1098.0000 | 1091.0000 | 1202.0000 | 1235.0000 | 1110.0000 | 1048.0000 | 1254.0000 | 1286.0000 | 1227.0000 | 1285.0000 | 1270.0000 | 1272.0000 | 1234.0000 | 1317.0000 | 1259.0000 | 1390.0000 | 1204.0000 | 1084.0000 | 1208.0000 | 1060.0000 | 1108.0000 | 1189.0000 | 1059.0000 | 1080.0000 | 1260.0000 | 1050.0000 | 1158.0000 | 1268.0000 | 963.0000 | 779.0000 | 697.0000 | 874.0000 | 1042.0000 | 787.0000 | 682.0000 | 698.0000 | 778.0000 | 940.6912 | 809.0000 | 810.0000 | 709.0000 | 657.0000 | 726.0000 | 738.0000 | 791.0000 | 551.0000 | 708.0000 | 983.0000 | 1043.0000 | 742.0000 | 958.0000 | |
| max | 2390.0000 | 2679.0000 | 2095.0000 | 2439.0000 | 2560.0000 | 2446.0000 | 2684.0000 | 2572.0000 | 2400.0000 | 2667.0000 | 2605.0000 | 2337.0000 | 2486.0000 | 2746.0000 | 2530.0000 | 2394.0000 | 2622.0000 | 2460.0000 | 2357.0000 | 2662.0000 | 2322.0000 | 2404.0000 | 2114.0000 | 1931.0000 | 2143.0000 | 2555.0000 | 2260.0000 | 2409.0000 | 2531.0000 | 2365.0000 | 2612.0000 | 2072.0000 | 2595.0000 | 2775.0000 | 2643.0000 | 2032.0000 | 2235.0000 | 2562.0000 | 2405.0000 | 2018.0000 | 2271.0000 | 1832.0000 | 1808.0000 | 1790.0000 | 1902.0000 | 2083.0000 | 1262.9826 | 1870.0000 | 1712.0000 | 1542.0000 | 1868.0000 | 1848.0000 | 1747.0000 | 2028.0000 | 2147.0000 | 2005.0000 | 1777.0000 | |
| mean | 1584.6275 | 1716.5119 | 1488.5952 | 1543.4471 | 1532.8688 | 1568.5417 | 1638.6488 | 1676.9583 | 1641.7798 | 1663.3690 | 1626.8929 | 1454.5900 | 1684.5952 | 1730.7083 | 1708.2599 | 1680.8475 | 1759.9167 | 1656.4405 | 1584.1905 | 1700.6190 | 1603.1298 | 1692.5893 | 1603.9775 | 1490.0774 | 1499.1718 | 1600.8512 | 1461.8185 | 1608.6488 | 1556.8214 | 1570.1988 | 1710.1369 | 1421.3036 | 1740.2381 | 1679.3393 | 1608.9405 | 1215.1548 | 1181.0238 | 1476.2976 | 1439.4702 | 1196.2619 | 1208.4137 | 1157.9733 | 1074.7370 | 1249.8472 | 1330.3214 | 1195.3155 | 879.7548 | 1051.5238 | 1088.2734 | 993.2143 | 1118.8452 | 959.4821 | 1116.2633 | 1292.9167 | 1420.1131 | 1144.0952 | 1195.0667 | |
| std | 253.7583 | 295.7079 | 213.8439 | 262.1349 | 244.7315 | 277.8202 | 307.7813 | 293.9112 | 246.4995 | 293.1831 | 264.1147 | 214.5271 | 214.0929 | 290.0779 | 221.7754 | 212.6782 | 258.9084 | 241.1909 | 217.6214 | 235.5518 | 183.9521 | 174.3749 | 164.8099 | 148.4322 | 139.3208 | 266.5683 | 175.2791 | 281.4597 | 349.5403 | 277.8914 | 291.0350 | 226.9915 | 301.4312 | 306.7480 | 366.4507 | 250.2112 | 310.9004 | 325.9123 | 288.2700 | 217.8347 | 231.8791 | 240.5562 | 170.6712 | 183.5437 | 268.1259 | 261.0409 | 126.4806 | 240.6183 | 213.9123 | 131.2727 | 197.5945 | 253.1675 | 228.1175 | 221.0499 | 237.9307 | 244.6727 | 245.5534 | |
| PT08.S5(O3) | count | 102.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 15.0000 |
| min | 384.0000 | 370.0000 | 344.0000 | 341.0000 | 320.0000 | 263.0000 | 305.0000 | 319.0000 | 343.0000 | 354.0000 | 360.0000 | 301.0000 | 445.0000 | 377.0000 | 334.0000 | 466.0000 | 427.0000 | 386.0000 | 437.0000 | 427.0000 | 393.0000 | 396.0000 | 404.0000 | 368.0000 | 456.0000 | 342.0000 | 385.0000 | 468.0000 | 382.0000 | 640.0000 | 549.0000 | 392.0000 | 568.0000 | 540.0000 | 440.0000 | 261.0000 | 296.0000 | 644.0000 | 483.0000 | 324.0000 | 387.0000 | 318.0000 | 337.0000 | 521.0000 | 463.0000 | 412.0000 | 253.0000 | 292.0000 | 337.0000 | 296.0000 | 348.0000 | 221.0000 | 262.0000 | 415.0000 | 544.0000 | 322.0000 | 489.0000 | |
| max | 1905.0000 | 2359.0000 | 1519.0000 | 1798.0000 | 1806.0000 | 2086.0000 | 1983.0000 | 2108.0000 | 1687.0000 | 1895.0000 | 2054.0000 | 1789.0000 | 2202.0000 | 2147.0000 | 1690.0000 | 1660.0000 | 2043.0000 | 2475.0000 | 1791.0000 | 2176.0000 | 1691.0000 | 1607.0000 | 1433.0000 | 1302.0000 | 1390.0000 | 1782.0000 | 2321.0000 | 1743.0000 | 1820.0000 | 2236.0000 | 2143.0000 | 1625.0000 | 2235.0000 | 2372.0000 | 2519.0000 | 1720.0000 | 2452.0000 | 2522.0000 | 2197.0000 | 2018.0000 | 2523.0000 | 2480.0000 | 2004.0000 | 2159.0000 | 1802.0000 | 2331.0000 | 1296.1468 | 2214.0000 | 2494.0000 | 1746.0000 | 1727.0000 | 1913.0000 | 2088.0000 | 2140.0000 | 2159.0000 | 1890.0000 | 1729.0000 | |
| mean | 1111.4706 | 1219.6607 | 862.0000 | 975.6502 | 894.4612 | 955.6071 | 1039.1667 | 1049.0298 | 860.2262 | 1040.2381 | 998.7798 | 829.3111 | 866.5833 | 967.9345 | 886.5883 | 917.1537 | 1024.4048 | 1044.0714 | 977.8631 | 1097.6310 | 851.5331 | 854.0179 | 753.6333 | 709.8869 | 759.3609 | 972.7976 | 993.2980 | 1025.3750 | 1007.5119 | 1187.7413 | 1241.5774 | 868.6845 | 1346.8155 | 1205.6429 | 1222.7083 | 855.0060 | 1117.5417 | 1503.7440 | 1140.2083 | 1026.1071 | 1151.6548 | 1224.1887 | 912.0268 | 1153.2352 | 1271.9345 | 1284.2560 | 630.2752 | 1212.5119 | 1144.6646 | 943.2202 | 920.8690 | 872.5417 | 1071.9753 | 1158.5714 | 1218.3333 | 788.1369 | 995.2667 | |
| std | 362.4545 | 395.3982 | 267.9472 | 332.4326 | 312.1580 | 386.0292 | 407.4759 | 393.8974 | 295.8590 | 349.9999 | 336.9261 | 288.3187 | 280.4743 | 348.4990 | 254.0365 | 267.5126 | 322.1283 | 436.7461 | 297.9020 | 384.9107 | 257.3851 | 244.5177 | 223.7792 | 195.1348 | 173.8073 | 325.0643 | 318.4881 | 311.1043 | 385.5823 | 349.1725 | 358.9085 | 278.1533 | 359.3962 | 369.6606 | 435.9667 | 351.1918 | 423.1223 | 446.3378 | 376.0609 | 318.1600 | 375.6491 | 532.3692 | 313.2340 | 327.9004 | 344.5220 | 476.6562 | 247.3312 | 457.0818 | 458.1082 | 345.3060 | 311.0878 | 445.5603 | 430.2276 | 384.8501 | 372.2695 | 330.6950 | 445.1219 | |
| T | count | 102.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 15.0000 |
| min | 6.1000 | 9.9000 | 6.3000 | 8.1000 | 7.4000 | 9.1000 | 9.5000 | 12.9000 | 7.4000 | 11.3000 | 14.9000 | 11.6000 | 16.4000 | 17.2000 | 18.3000 | 17.8000 | 20.2000 | 19.7000 | 16.2000 | 21.2000 | 20.2000 | 20.7000 | 22.4000 | 18.6000 | 19.5000 | 19.7000 | 19.1849 | 16.8000 | 12.2000 | 11.9000 | 17.1000 | 13.9000 | 12.6000 | 14.6000 | 9.9000 | 3.2000 | 1.3000 | 2.6000 | 8.3000 | 4.0000 | 2.2000 | 1.2000 | 2.1000 | 5.4000 | 2.4000 | 1.8000 | 1.0000 | 0.3000 | 2.9000 | 0.6000 | 1.4000 | -1.9000 | 0.3000 | 7.2000 | 10.3000 | 8.9000 | 9.5000 | |
| max | 22.2000 | 29.3000 | 21.3000 | 27.6000 | 22.7000 | 21.5000 | 31.3000 | 28.4000 | 30.9000 | 31.5000 | 32.8000 | 32.6000 | 31.8000 | 40.6000 | 37.6000 | 42.2000 | 40.4000 | 42.6000 | 41.4000 | 44.6000 | 40.4000 | 39.3000 | 40.5000 | 39.9000 | 38.3000 | 40.7000 | 38.4000 | 35.2000 | 34.3000 | 28.9000 | 30.8000 | 24.4000 | 27.2000 | 24.9000 | 26.8000 | 21.5000 | 18.8000 | 16.8000 | 19.2000 | 20.3000 | 16.8000 | 14.9000 | 15.0000 | 16.9000 | 15.8000 | 15.8000 | 11.8000 | 15.7000 | 19.9000 | 13.8000 | 13.0000 | 14.8000 | 20.2000 | 28.8000 | 25.7000 | 30.0000 | 28.5000 | |
| mean | 12.4735 | 17.1143 | 12.9298 | 16.3741 | 15.5247 | 14.4048 | 18.1363 | 19.3083 | 16.4000 | 19.5726 | 22.8036 | 21.8494 | 22.7887 | 27.0565 | 25.8681 | 27.5034 | 30.3208 | 29.4786 | 27.4411 | 31.1625 | 29.6378 | 28.8214 | 30.3258 | 29.6411 | 28.0161 | 28.5601 | 26.7196 | 23.7685 | 22.5405 | 21.3071 | 23.0690 | 17.7946 | 20.9518 | 19.2679 | 19.6476 | 11.6560 | 11.8976 | 10.7708 | 13.1792 | 13.8345 | 10.1093 | 8.2755 | 9.5650 | 10.5553 | 10.0161 | 7.8143 | 4.9785 | 6.7310 | 8.2445 | 8.1381 | 5.8982 | 5.3387 | 10.5354 | 15.8310 | 17.4226 | 16.9470 | 16.9600 | |
| std | 4.1068 | 4.5795 | 3.7026 | 4.5561 | 3.8951 | 2.9525 | 5.8160 | 4.0352 | 3.7253 | 4.6115 | 4.9091 | 5.2302 | 3.9992 | 6.5377 | 4.9064 | 5.8770 | 5.5971 | 6.0386 | 6.5469 | 6.3539 | 5.4067 | 5.0966 | 4.8973 | 5.2503 | 5.0646 | 5.0561 | 4.4427 | 4.0305 | 5.0300 | 3.8758 | 3.4857 | 2.6777 | 3.4374 | 2.4058 | 3.3871 | 3.7562 | 4.0488 | 3.3541 | 2.4367 | 3.1609 | 3.0726 | 2.8332 | 2.9209 | 3.2415 | 3.3265 | 3.2456 | 2.0677 | 3.9796 | 3.4173 | 2.7299 | 2.3896 | 3.8535 | 5.1708 | 5.4188 | 3.6192 | 4.5581 | 7.0894 | |
| RH | count | 102.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 15.0000 |
| min | 28.4000 | 14.9000 | 23.1000 | 20.0000 | 18.9000 | 27.3000 | 16.2000 | 20.4000 | 19.7000 | 19.0000 | 17.1000 | 9.2000 | 22.3000 | 14.2000 | 18.8000 | 11.9000 | 12.7000 | 9.6000 | 11.8000 | 12.3000 | 13.7000 | 21.1000 | 15.9000 | 10.0000 | 15.4000 | 16.0000 | 13.8000 | 22.4000 | 23.0000 | 21.3000 | 31.8000 | 35.2000 | 43.0000 | 47.1000 | 32.1000 | 31.3000 | 14.0000 | 36.7000 | 49.1000 | 18.9000 | 16.3000 | 29.6000 | 27.5000 | 45.6798 | 30.8000 | 20.8000 | 25.9000 | 17.2000 | 20.0000 | 18.0000 | 30.6000 | 13.5000 | 14.4000 | 15.1000 | 22.8000 | 9.9000 | 13.1000 | |
| max | 81.1000 | 71.1000 | 83.2000 | 78.1000 | 80.6000 | 82.4000 | 80.5000 | 78.6000 | 85.2000 | 70.5000 | 61.6000 | 54.8000 | 82.3000 | 62.3000 | 60.8000 | 60.9000 | 59.6000 | 53.2000 | 59.8000 | 69.3000 | 63.6000 | 81.8000 | 68.0000 | 67.4000 | 66.9000 | 66.8000 | 70.4000 | 87.2000 | 76.6000 | 71.8000 | 80.5000 | 84.6000 | 78.4000 | 86.5000 | 78.1000 | 87.1000 | 76.7000 | 85.7000 | 88.7000 | 81.4000 | 84.5000 | 82.9000 | 83.5000 | 83.8000 | 86.6000 | 83.8000 | 80.4000 | 74.5000 | 85.0000 | 70.0000 | 86.6000 | 84.0000 | 80.9000 | 74.5000 | 83.6000 | 80.6000 | 63.1000 | |
| mean | 53.9480 | 48.9935 | 52.3917 | 45.9064 | 49.8732 | 57.4810 | 48.2167 | 48.0571 | 59.5018 | 44.6732 | 35.8518 | 32.5610 | 48.6970 | 36.3637 | 39.9022 | 35.9341 | 35.6970 | 31.2583 | 31.9554 | 34.7726 | 34.3085 | 48.3190 | 44.1586 | 37.7554 | 39.9565 | 38.8024 | 36.6985 | 52.3054 | 47.8577 | 50.7937 | 56.7125 | 59.5875 | 63.0071 | 69.9976 | 60.8851 | 60.4292 | 43.8536 | 69.0298 | 73.8655 | 50.9387 | 56.7870 | 54.9694 | 54.1148 | 61.9274 | 65.3905 | 55.8911 | 46.5545 | 44.9762 | 49.3517 | 43.1405 | 68.3095 | 47.8964 | 45.8875 | 48.8429 | 56.3845 | 47.0440 | 42.7867 | |
| std | 12.7451 | 14.0011 | 14.3069 | 14.3195 | 15.0211 | 14.0707 | 17.8368 | 15.0541 | 13.8906 | 14.0491 | 11.8934 | 12.7115 | 16.4059 | 12.8636 | 10.7822 | 12.9813 | 12.3616 | 11.9905 | 12.7920 | 13.5396 | 12.6837 | 15.3796 | 15.0325 | 12.3751 | 12.7067 | 14.0423 | 12.6528 | 14.9754 | 13.2462 | 12.4752 | 11.2547 | 12.6154 | 8.7083 | 8.1340 | 11.6663 | 13.7894 | 15.4792 | 11.4762 | 9.0333 | 16.5186 | 14.6960 | 14.9442 | 13.6297 | 8.5666 | 11.8126 | 15.2033 | 8.3198 | 14.3491 | 13.3080 | 11.6875 | 14.6919 | 17.2195 | 14.8886 | 16.3214 | 15.2100 | 17.9383 | 18.5104 | |
| AH | count | 102.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 168.0000 | 15.0000 |
| min | 0.6195 | 0.5237 | 0.5319 | 0.4023 | 0.4969 | 0.4868 | 0.5625 | 0.6760 | 0.6445 | 0.6686 | 0.5673 | 0.3754 | 0.8811 | 0.7502 | 1.0172 | 0.7613 | 0.7749 | 0.7526 | 0.7158 | 0.9941 | 0.8335 | 1.3863 | 1.0296 | 0.6591 | 0.9692 | 0.8162 | 0.8342 | 1.0272 | 0.6514 | 0.7773 | 1.3546 | 0.9107 | 1.0581 | 1.3156 | 0.6073 | 0.4329 | 0.1988 | 0.4412 | 0.8674 | 0.3119 | 0.2749 | 0.3340 | 0.3766 | 0.5948 | 0.4100 | 0.3422 | 0.2726 | 0.2477 | 0.3033 | 0.2269 | 0.3962 | 0.1847 | 0.2672 | 0.5782 | 0.6346 | 0.3866 | 0.5028 | |
| max | 0.9341 | 1.0945 | 1.0919 | 1.0912 | 1.0717 | 1.2695 | 1.2953 | 1.4852 | 1.4681 | 1.3250 | 1.2771 | 1.2148 | 1.7040 | 1.5020 | 1.4733 | 1.7193 | 1.9390 | 1.6308 | 1.4452 | 2.0042 | 1.9998 | 2.1806 | 2.1719 | 2.0767 | 1.8710 | 1.6872 | 2.2310 | 1.9776 | 1.7261 | 1.6261 | 1.8881 | 1.6384 | 1.8632 | 2.0224 | 1.9800 | 1.1328 | 1.1455 | 1.2700 | 1.3713 | 1.2326 | 1.1106 | 1.1381 | 1.0851 | 0.9971 | 1.0567 | 0.9972 | 0.6791 | 0.6967 | 1.0859 | 0.7413 | 0.7530 | 0.7382 | 0.9292 | 1.0250 | 1.3930 | 1.2839 | 0.8275 | |
| mean | 0.7558 | 0.9008 | 0.7516 | 0.8269 | 0.8399 | 0.9236 | 0.9267 | 1.0292 | 1.0755 | 0.9556 | 0.9383 | 0.7907 | 1.2721 | 1.1944 | 1.2644 | 1.2181 | 1.4338 | 1.1711 | 1.0504 | 1.4228 | 1.3310 | 1.7798 | 1.7734 | 1.4780 | 1.4320 | 1.3998 | 1.2124 | 1.4728 | 1.2768 | 1.2603 | 1.5467 | 1.1920 | 1.5390 | 1.5401 | 1.3835 | 0.8282 | 0.6146 | 0.9023 | 1.1123 | 0.8001 | 0.7107 | 0.6257 | 0.6508 | 0.8033 | 0.8090 | 0.5868 | 0.4105 | 0.4295 | 0.5414 | 0.4605 | 0.6275 | 0.4341 | 0.5751 | 0.8146 | 1.0775 | 0.8645 | 0.7248 | |
| std | 0.0816 | 0.0972 | 0.1143 | 0.1888 | 0.1364 | 0.1830 | 0.1403 | 0.2108 | 0.1799 | 0.1343 | 0.1633 | 0.1898 | 0.2195 | 0.1632 | 0.0996 | 0.2033 | 0.2492 | 0.1713 | 0.1233 | 0.2065 | 0.2844 | 0.1415 | 0.2711 | 0.3262 | 0.2233 | 0.2041 | 0.2565 | 0.2749 | 0.3113 | 0.3013 | 0.1229 | 0.2121 | 0.2014 | 0.1303 | 0.2870 | 0.2055 | 0.2587 | 0.2060 | 0.1315 | 0.2698 | 0.1723 | 0.2488 | 0.1930 | 0.1400 | 0.1790 | 0.1513 | 0.0829 | 0.0975 | 0.1655 | 0.1044 | 0.0906 | 0.1656 | 0.1824 | 0.1030 | 0.1740 | 0.2566 | 0.0978 |
Data Extraction
Description for attributes
- Date (DD/MM/YYYY)
- Time (HH.MM.SS)
- [CO(GT)]: True hourly averaged concentration CO in mg/m^3 (reference analyzer)
- [PT08.S1]: (tin oxide) hourly averaged sensor response (nominally CO targeted)
- [NMHC(GT)]: True hourly averaged overall Non Metanic HydroCarbons concentration in microg/m^3 (reference analyzer)
- [C6H6(GT)]: True hourly averaged Benzene concentration in microg/m^3 (reference analyzer)
- [PT08.S2]: (titania) hourly averaged sensor response (nominally NMHC targeted)
- [NOx(GT)]: True hourly averaged NOx concentration in ppb (reference analyzer)
- [PT08.S3]: (tungsten oxide) hourly averaged sensor response (nominally NOx targeted)
- [NO2(GT)]: True hourly averaged NO2 concentration in microg/m^3 (reference analyzer)
- [PT08.S4]: (tungsten oxide) hourly averaged sensor response (nominally NO2 targeted)
- [PT08.S5]: (indium oxide) hourly averaged sensor response (nominally O3 targeted)
- [T]: Temperature in °C
- [RH]: Relative Humidity (%)
- [AH]: Absolute Humidity
Data Interpolation
Data description after interpolation through Gaussian Process(GP)


cross-sectional description
| column | count | normality | l_shift | r_shift | iqr_min | iqr_25 | mean | iqr_75 | iqr_max | std | diff_maxmin | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | CO(GT) | 9357.0 | False | True | False | 0.1000 | 1.1000 | 2.102615 | 2.800000 | 11.900 | 1.371028 | 11.8000 |
| 1 | PT08.S1(CO) | 9357.0 | False | True | False | 647.0000 | 938.0000 | 1097.691746 | 1222.000000 | 2040.000 | 213.893057 | 1393.0000 |
| 2 | NMHC(GT) | 1231.0 | False | True | False | 7.0000 | 76.0000 | 218.553901 | 297.000000 | 1189.000 | 184.403001 | 1182.0000 |
| 3 | C6H6(GT) | 9357.0 | False | True | False | 0.1000 | 4.5000 | 10.033281 | 13.700000 | 63.700 | 7.336185 | 63.6000 |
| 4 | PT08.S2(NMHC) | 9357.0 | False | True | False | 383.0000 | 736.0000 | 936.686728 | 1105.000000 | 2214.000 | 263.138532 | 1831.0000 |
| 5 | NOx(GT) | 9357.0 | False | True | False | 2.0000 | 95.0000 | 237.567906 | 322.000000 | 1479.000 | 202.292320 | 1477.0000 |
| 6 | PT08.S3(NOx) | 9357.0 | False | True | False | 322.0000 | 664.0000 | 834.655380 | 965.000000 | 2683.000 | 252.980266 | 2361.0000 |
| 7 | NO2(GT) | 9357.0 | False | True | False | 2.0000 | 73.0000 | 108.064926 | 135.000000 | 340.000 | 46.594004 | 338.0000 |
| 8 | PT08.S4(NO2) | 9357.0 | False | False | True | 551.0000 | 1216.0000 | 1450.059221 | 1665.000000 | 2775.000 | 344.348153 | 2224.0000 |
| 9 | PT08.S5(O3) | 9357.0 | False | True | False | 221.0000 | 737.0000 | 1021.100171 | 1257.000000 | 2523.000 | 392.316597 | 2302.0000 |
| 10 | T | 9357.0 | False | True | False | -1.9000 | 11.5000 | 18.186668 | 24.300000 | 44.600 | 8.851993 | 46.5000 |
| 11 | RH | 9357.0 | False | False | True | 9.2000 | 36.2000 | 49.220253 | 61.935725 | 88.700 | 17.093713 | 79.5000 |
| 12 | AH | 9357.0 | False | True | False | 0.1847 | 0.7288 | 1.019927 | 1.306700 | 2.231 | 0.403081 | 2.0463 |

longitudinal description
| Normality | Heteroskedasticity | Autocorrelation | Stationary | Summary | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| jb_stats | jb_pval | skew | kurtosis | determination | bh_stats | bh_pval | determination | lb_stats | lb_pval | determination | adf_stats | adf_pval | adf_used_lag | adf_nobs | adf_critical_value_5% | kpss_stats | kpss_pval | kpss_used_lag | kpss_critical_values_5% | adf_determination | kpss_determination | determination | Autocorrelation | Heteroskedasticity | Normality | Stationary | |
| CO(GT) | 7006.070249 | 0.000000e+00 | 1.427208 | 6.134064 | False | 1.319321 | 1.116028e-14 | True | 6672.912750 | 0.000000e+00 | True | -10.267951 | 4.072187e-18 | 37 | 9319 | -2.861850 | 1.326229 | 0.010000 | 42 | 0.463 | True | False | False | True | True | False | False |
| PT08.S1(CO) | 1028.166592 | 5.450814e-224 | 0.781977 | 3.437283 | False | 1.011468 | 7.501991e-01 | False | 7317.908522 | 0.000000e+00 | True | -9.701349 | 1.070125e-16 | 38 | 9318 | -2.861850 | 0.491353 | 0.043614 | 47 | 0.463 | True | False | False | True | False | False | False |
| C6H6(GT) | 5729.397567 | 0.000000e+00 | 1.385777 | 5.648389 | False | 0.799953 | 4.792266e-10 | True | 6584.589871 | 0.000000e+00 | True | -10.300587 | 3.380466e-18 | 37 | 9319 | -2.861850 | 0.830241 | 0.010000 | 42 | 0.463 | True | False | False | True | True | False | False |
| PT08.S2(NMHC) | 535.960078 | 4.147132e-117 | 0.581900 | 3.142361 | False | 0.874264 | 1.764749e-04 | True | 7051.977577 | 0.000000e+00 | True | -10.395797 | 1.966836e-18 | 37 | 9319 | -2.861850 | 1.042781 | 0.010000 | 43 | 0.463 | True | False | False | True | True | False | False |
| NOx(GT) | 10697.346443 | 0.000000e+00 | 1.765343 | 6.869386 | False | 7.652051 | 0.000000e+00 | True | 7695.744176 | 0.000000e+00 | True | -6.883601 | 1.412917e-09 | 26 | 9330 | -2.861850 | 9.428993 | 0.010000 | 50 | 0.463 | True | False | False | True | True | False | False |
| PT08.S3(NOx) | 5037.073665 | 0.000000e+00 | 1.118870 | 5.812869 | False | 0.758071 | 1.150333e-14 | True | 7695.240985 | 0.000000e+00 | True | -10.581876 | 6.872173e-19 | 36 | 9320 | -2.861850 | 3.567417 | 0.010000 | 49 | 0.463 | True | False | False | True | True | False | False |
| NO2(GT) | 1127.862309 | 1.224085e-245 | 0.764447 | 3.745229 | False | 2.018402 | 5.928394e-84 | True | 7619.229219 | 0.000000e+00 | True | -7.526296 | 3.677213e-11 | 32 | 9324 | -2.861850 | 8.069080 | 0.010000 | 48 | 0.463 | True | False | False | True | True | False | False |
| PT08.S4(NO2) | 82.452356 | 1.246517e-18 | 0.226822 | 3.075445 | False | 0.528635 | 9.887155e-70 | True | 7797.407995 | 0.000000e+00 | True | -5.966929 | 1.977626e-07 | 37 | 9319 | -2.861850 | 9.684327 | 0.010000 | 53 | 0.463 | True | False | False | True | True | False | False |
| PT08.S5(O3) | 653.751364 | 1.095708e-142 | 0.642244 | 3.164052 | False | 1.246734 | 7.685124e-10 | True | 7563.208040 | 0.000000e+00 | True | -11.032393 | 5.625545e-20 | 36 | 9320 | -2.861850 | 1.065825 | 0.010000 | 48 | 0.463 | True | False | False | True | True | False | False |
| T | 254.963898 | 4.318104e-56 | 0.323656 | 2.515282 | False | 0.264708 | 1.000786e-281 | True | 9124.195826 | 0.000000e+00 | True | -2.883684 | 4.727850e-02 | 29 | 9327 | -2.861850 | 7.578053 | 0.010000 | 55 | 0.463 | True | False | False | True | True | False | False |
| RH | 238.984833 | 1.273805e-52 | -0.042656 | 2.221733 | False | 1.471940 | 5.276172e-27 | True | 8692.293601 | 0.000000e+00 | True | -7.329900 | 1.135061e-10 | 38 | 9318 | -2.861850 | 3.128120 | 0.010000 | 52 | 0.463 | True | False | False | True | True | False | False |
| AH | 243.143172 | 1.592689e-53 | 0.270941 | 2.425537 | False | 0.497165 | 3.687968e-83 | True | 9216.839755 | 0.000000e+00 | True | -5.069178 | 1.616381e-05 | 25 | 9331 | -2.861850 | 4.324071 | 0.010000 | 57 | 0.463 | True | False | False | True | True | False | False |
| NMHC(GT) | 955.878225 | 2.714443e-208 | 1.571748 | 5.958812 | False | 2.448508 | 4.847462e-19 | True | 765.148931 | 2.039492e-168 | True | -2.937991 | 4.110514e-02 | 23 | 1207 | -2.863938 | 1.176806 | 0.010000 | 16 | 0.463 | True | False | False | True | True | False | False |
Missing Value & Duplication Inspection
The number of total data is 9,357 and number of datum with missing values was counted monthly in the data.

before interpolation
| column | total | missing-value | duplication | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| quasi-dtypes | freq | not_freq | freq | ratio | rank | cardinality | selectivity | rank | ||
| 0 | CO(GT) | numeric | 9357 | 7674 | 1683 | 0.179865 | 2.0 | 97 | 0.010367 | 1.0 |
| 1 | PT08.S1(CO) | numeric | 9357 | 8991 | 366 | 0.039115 | 9.0 | 1042 | 0.111360 | 8.0 |
| 2 | NMHC(GT) | numeric | 9357 | 914 | 8443 | 0.902319 | 1.0 | 430 | 0.045955 | 4.0 |
| 3 | C6H6(GT) | numeric | 9357 | 8991 | 366 | 0.039115 | 9.0 | 408 | 0.043604 | 3.0 |
| 4 | PT08.S2(NMHC) | numeric | 9357 | 8991 | 366 | 0.039115 | 9.0 | 1246 | 0.133162 | 10.0 |
| 5 | NOx(GT) | numeric | 9357 | 7718 | 1639 | 0.175163 | 4.0 | 926 | 0.098963 | 7.0 |
| 6 | PT08.S3(NOx) | numeric | 9357 | 8991 | 366 | 0.039115 | 9.0 | 1222 | 0.130597 | 9.0 |
| 7 | NO2(GT) | numeric | 9357 | 7715 | 1642 | 0.175484 | 3.0 | 284 | 0.030352 | 2.0 |
| 8 | PT08.S4(NO2) | numeric | 9357 | 8991 | 366 | 0.039115 | 9.0 | 1604 | 0.171422 | 11.0 |
| 9 | PT08.S5(O3) | numeric | 9357 | 8991 | 366 | 0.039115 | 9.0 | 1744 | 0.186385 | 12.0 |
| 10 | T | numeric | 9357 | 8991 | 366 | 0.039115 | 9.0 | 437 | 0.046703 | 5.0 |
| 11 | RH | numeric | 9357 | 8991 | 366 | 0.039115 | 9.0 | 754 | 0.080581 | 6.0 |
| 12 | AH | numeric | 9357 | 8991 | 366 | 0.039115 | 9.0 | 6684 | 0.714332 | 13.0 |
after interpolation
| column | total | missing-value | duplication | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| quasi-dtypes | freq | not_freq | freq | ratio | rank | cardinality | selectivity | rank | ||
| 0 | CO(GT) | numeric | 9357 | 9357 | 0 | 0.000000 | 7.5 | 1779 | 0.190125 | 8.0 |
| 1 | PT08.S1(CO) | numeric | 9357 | 9357 | 0 | 0.000000 | 7.5 | 1406 | 0.150262 | 5.0 |
| 2 | NMHC(GT) | numeric | 9357 | 1231 | 8126 | 0.868441 | 1.0 | 747 | 0.079833 | 1.0 |
| 3 | C6H6(GT) | numeric | 9357 | 9357 | 0 | 0.000000 | 7.5 | 772 | 0.082505 | 2.0 |
| 4 | PT08.S2(NMHC) | numeric | 9357 | 9357 | 0 | 0.000000 | 7.5 | 1611 | 0.172171 | 7.0 |
| 5 | NOx(GT) | numeric | 9357 | 9357 | 0 | 0.000000 | 7.5 | 2564 | 0.274019 | 12.0 |
| 6 | PT08.S3(NOx) | numeric | 9357 | 9357 | 0 | 0.000000 | 7.5 | 1587 | 0.169606 | 6.0 |
| 7 | NO2(GT) | numeric | 9357 | 9357 | 0 | 0.000000 | 7.5 | 1925 | 0.205728 | 9.0 |
| 8 | PT08.S4(NO2) | numeric | 9357 | 9357 | 0 | 0.000000 | 7.5 | 1969 | 0.210431 | 10.0 |
| 9 | PT08.S5(O3) | numeric | 9357 | 9357 | 0 | 0.000000 | 7.5 | 2109 | 0.225393 | 11.0 |
| 10 | T | numeric | 9357 | 9357 | 0 | 0.000000 | 7.5 | 802 | 0.085711 | 3.0 |
| 11 | RH | numeric | 9357 | 9357 | 0 | 0.000000 | 7.5 | 1119 | 0.119590 | 4.0 |
| 12 | AH | numeric | 9357 | 9357 | 0 | 0.000000 | 7.5 | 7049 | 0.753340 | 13.0 |
References
'quantitative analysis > analysis report' 카테고리의 다른 글
| [Classification] Bank Marketing (1) | 2023.05.07 |
|---|